
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Roxana Pincheira | + 4491 word(s) | 4491 | 2021-12-20 05:15:46 | | | |
| 2 | Peter Tang | -17 word(s) | 4474 | 2022-01-11 07:36:30 | | |
Video Upload Options
SALL proteins are a family of four conserved C2H2 zinc finger transcription factors that play critical roles in organogenesis during embryonic development. They regulate cell proliferation, survival, migration, and stemness; consequently, they are involved in various human genetic disorders and cancer. SALL4 is a well-recognized oncogene; however, SALL1–3 play dual roles depending on the cancer context and stage of the disease.
1. Introduction
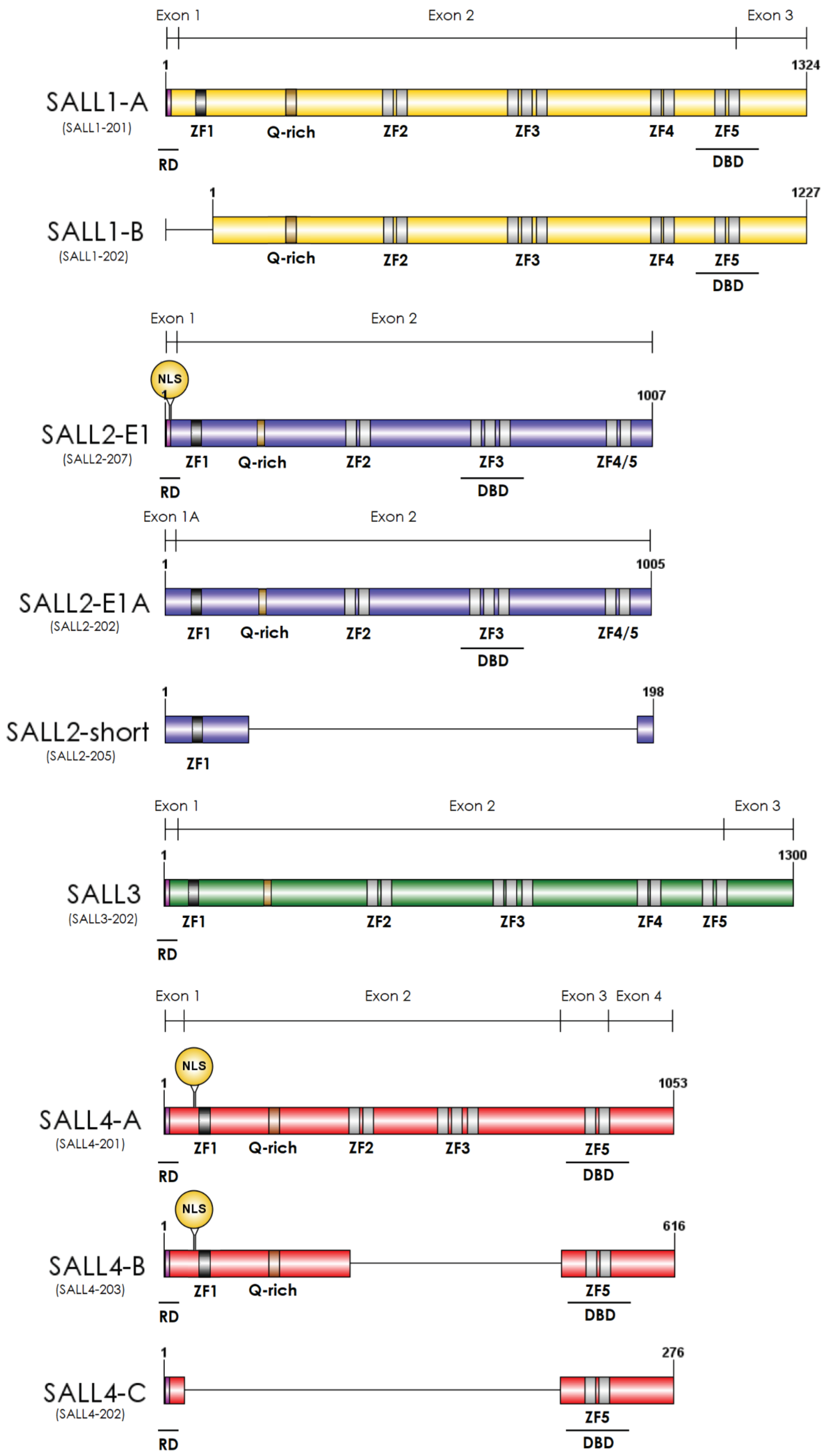
2. Essential Roles of SALL Genes during Development
3. Common Cellular Functions and Targets of the SALL Proteins in Cancer
3.1. Cell Proliferation
3.2. Apoptosis and Cell Survival
3.3. Cell Migration and Invasion
3.4. Stemness
4. Common Regulatory Mechanisms for SALL Proteins in Cancer
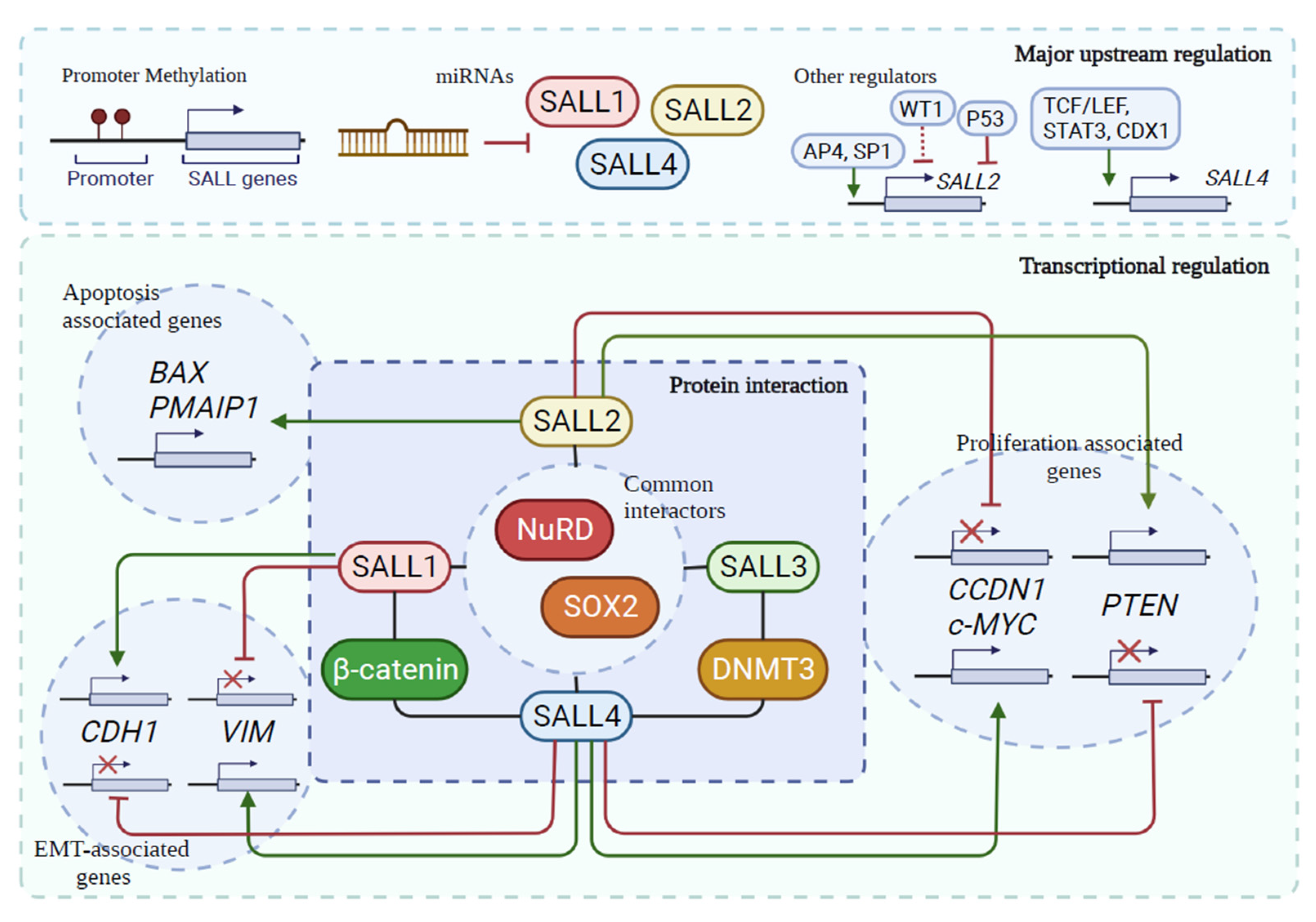
|
Cancer Type/Cellular Model |
microRNA |
Target |
SALL Status/Key Findings |
Experimental Approach |
Ref. |
|---|---|---|---|---|---|
|
Glioma/Glioblastoma |
miR-302/367 cluster |
SALL2 |
miR-302/367 cluster can reprogram tumor cells, generating more benign phenotypes by suppressing OCT3/4, SOX2, KLF4, c-MYC, POU3F2, OLIG2, and SALL2 |
qRT-PCR, cytokine array analysis |
[94] |
|
Glioma/Glioblastoma |
miR-16 |
SALL4 |
miR-16 inhibits proliferation, migration, and invasion in glioma cells by directly targeting SALL4 |
qRT-PCR and Luciferase reporter assay |
[95] |
|
Glioma/Glioblastoma |
miR-103/miR-195/miR-15-B |
SALL4 |
miR-103, miR-195, and miR-15-B inhibit proliferation, migration, and invasion and promote apoptosis in glioma by directly targeting SALL4 |
qRT-PCR, Western blot, and Luciferase reporter assay |
[96] |
|
Glioma/Glioblastoma |
miR-107 |
SALL4 |
miR-107 inhibits proliferation and promotes apoptosis in glioma cells by directly targeting SALL4 |
qRT-PCR, Western blot, and Luciferase reporter assay |
[97] |
|
Glioma/Glioblastoma |
miR-181b |
SALL4 |
miR-181b inhibits proliferation, migration, and invasion and promotes apoptosis in glioma by directly targeting SALL4 |
qRT-PCR, Western blot, and Luciferase reporter assay |
[98] |
|
Gastric cancer |
miR188-5p |
SALL4 |
miR-188-5p promotes proliferation and migration by upregulating SALL4 expression, nuclear translocation, and transcription |
qRT-PCR, Western blot, and Luciferase reporter assay |
[99] |
|
Gastric cancer |
miR-16 |
SALL4 |
miR-16 inhibits proliferation and migration in GC by directly targeting SALL4 |
qRT-PCR and Luciferase reporter assay |
[100] |
|
Colorectal cancer |
miR-181a-2 * |
SALL1 |
miR-181a-2 * correlates with a trend of repression of SALL1 and high methylation status of the SALL1 promoter |
qRT-PCR and bisulfite modification followed by quantitative methylation- specific PCR (qMSP) |
[101] |
|
Colorectal cancer |
miR-219-5p |
SALL4 |
miR-219-5p inhibits proliferation, migration, and invasion, reduces drug resistance, and promotes apoptosis in CRC by directly targeting SALL4 |
qRT-PCR, Western blot, and Luciferase reporter assay |
[102] |
|
Colorectal cancer |
miR-3622a-3p |
SALL4 |
miR-3622a-3p inhibits proliferation, cell cycle, migration, invasion, and stemness features and promotes apoptosis by targeting SALL4 |
qRT-PCR, Luciferase assay, RNA immunoprecipitation (RIP) assay, and pull-down assay |
[103] |
|
Embryonic stem cell |
miR15-B |
SALL4 |
Anti-miR-15b enhances expansion of HSC in vitro by targeting SALL4 |
qRT-PCR |
[104] |
|
Embryonic stem cell |
miR-294 and let-7 miRNAs |
SALL4 |
Let-7 miR family inhibits self-renewal genes, and miR-294 indirectly induces self-renewal genes, including SALL4 |
qRT-PCR, Western blot, and Luciferase reporter assay |
[105] |
|
Oral squamous cell carcinoma |
miR-103 |
SALL4 |
miR-103 inhibits cell proliferation and invasion by downregulating SALL4 mRNA in Tca8113 cells |
Luciferase reporter assay |
[106] |
|
Breast cancer |
SNHG12 and miR-15a-5p |
SALL4 |
Long non-coding RNA (lncRNA) small nucleolar RNA host gene 12 (SNHG12) promotes proliferation, migration, and invasion and inhibits apoptosis in breast cancer by upregulating SALL4 expression via sponging miR-15a-5p; SALL4 is a direct target of miR-15a-5p |
qRT-PCR, Western blot, and Luciferase reporter assay |
[107] |
|
Renal cell carcinoma |
miR-942 |
SALL1 |
miR-942 affects the survival of patients with renal cell carcinoma by negatively regulating the expression of SALL1 |
RNA-seq and qRT-PCR |
[108] |
|
Prostate cancer |
miR-4286 |
SALL1 |
miR-4286 regulates proliferation and apoptosis in PCa cells by directly targeting the 3′UTR of SALL1 mRNA |
qRT-PCR and Luciferase reporter assay |
[109] |
|
Lung cancer |
HOXA11-AS and miR-3619-5p |
SALL4 |
lncRNA homeobox A11 antisense (HOXA11-AS) promotes proliferation, migration, invasion, and glycolysis in non-small cell lung cancer (NSCLC) cells by upregulating SALL4 expression via sponging miR-3619-5p; SALL4 is a direct target of miR-3619-5p |
qRT-PCR, Western blot, and Luciferase reporter assay |
[110] |
|
Osteosarcoma |
ZEB2-AS1 and miR-107 |
SALL4 |
lncRNA ZEB2-AS1 (ZEB2-AS1) promotes proliferation, invasion, and metastasis and inhibits apoptosis in osteosarcoma cells by upregulating SALL4 expression via sponging miR-107; SALL4 is a direct target of miR-107 |
qRT-PCR, Luciferase assay, and RNA pull-down assay |
[111] |
|
Hepatocellular carcinoma |
miR-296-5p |
SALL4 |
miR-296-5p inhibits stemness potency of hepatocellular carcinoma (HCC) cells via the Brg1/Sall4 axis; Brg1 binds to the SALL4 promoter |
qRT-PCR, Western blot, Luciferase reporter assay, and Chromatin immunoprecipitation (ChIP) assay |
[112] |
|
Hepatocellular carcinoma |
miR-15a |
SALL4 |
Exosomal miR-15a reduces proliferation, migration, invasion, and survival by directly targeting SALL4 |
qRT-PCR, Western blot, and Luciferase reporter assay |
[113] |
5. SALL Proteins in Cancer
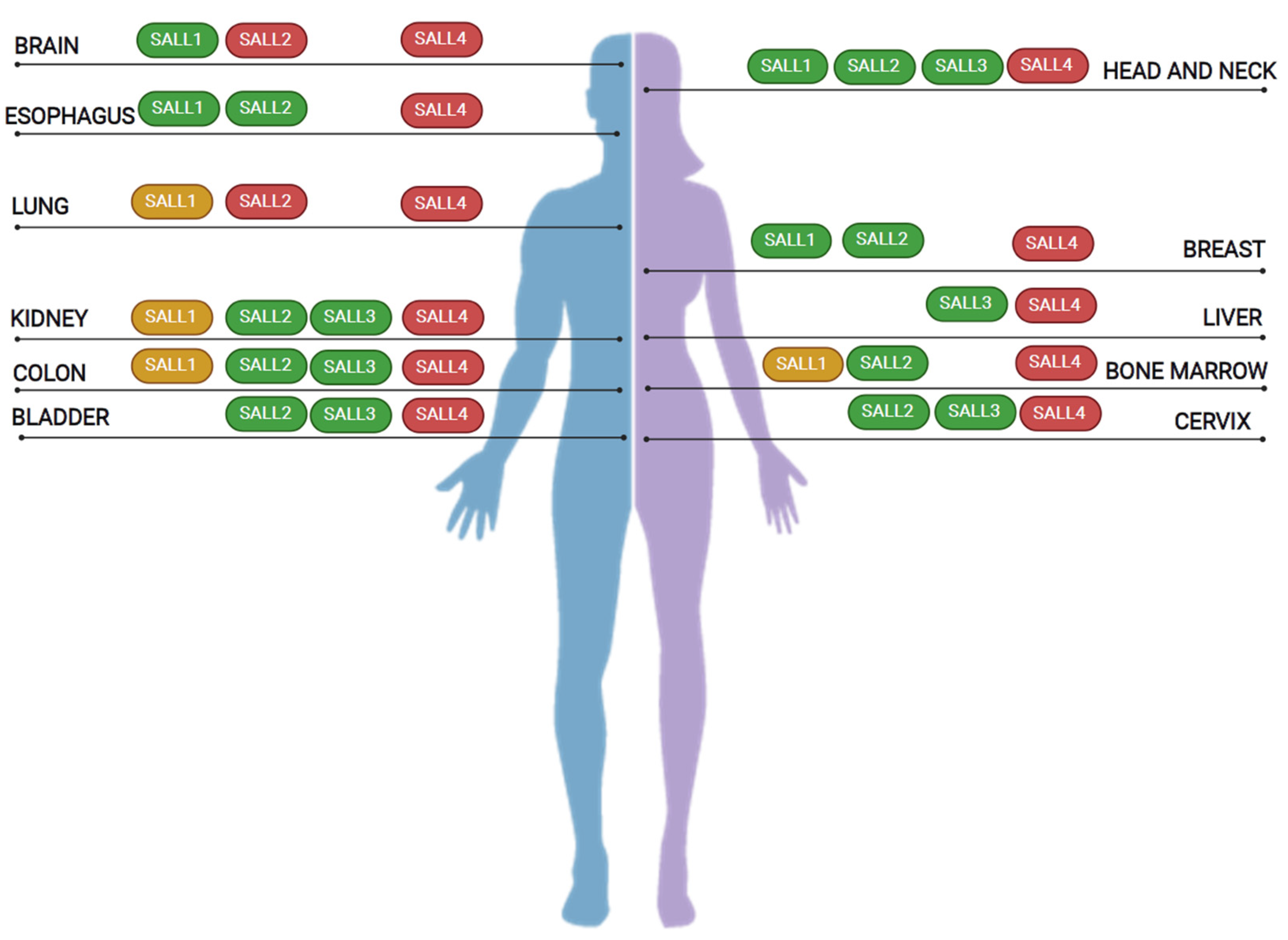
|
Cancer Type |
SALL Member |
Expression Levels |
Genetic Alteration/Regulation |
Association With Cancer/Biological Process |
Proposed Cancer Role |
Ref. |
|---|---|---|---|---|---|---|
|
Lung |
SALL1 |
High |
Undescribed |
Expression correlated with lower overall survival of NSCLC patients |
Oncogene |
[115] |
|
Lung |
SALL2 |
Low |
LOH |
Undescribed |
Undescribed |
[64] |
|
Lung |
SALL4 |
High |
Undescribed |
Expressed in 88% of the lung cancer samples May be used as a diagnostic marker |
Oncogene |
[116] |
|
Lung |
SALL4 |
High |
Undescribed |
SALL4 knockdown inhibits cell proliferation by cell cycle arrest at the GO/G1 phase Loss of SALL4 function inhibits migration, invasion and reduces the size of the transplanted tumor in an in vivo model. |
Oncogene |
[34] |
|
Lung |
SALL4 |
High |
Undescribed |
SALL4 silencing sensitizes cells to cisplatin, carboplatin, and paclitaxel treatment |
Oncogene |
[117] |
|
Esophageal |
SALL1 |
Low |
Hypermethylation |
SALL1, ADHFE1, EOMES, and TFPI2 are proposed as part of a tumor suppressors panel with diagnostic relevance |
Tumor suppressor |
|
|
Esophageal |
SALL2 |
Low in radioresistant ESCC cell lines |
Hypermethylation |
SALL2 overexpression enhances apoptosis after radiation and decreases migration, viability, and cisplatin resistance in TE-1/R and Eca-109/R cell lines |
Tumor suppressor |
[48] |
|
Esophageal |
SALL4 |
High |
Undescribed |
SALL4 silencing in ESCC cells is associated with suppressing cell migration, invasion, viability, and drug resistance in vivo SALL4 knockdown reduces epithelial-mesenchymal transition by targeting the Wnt/β-catenin signaling pathway |
Oncogene |
|
|
Bladder |
SALL2 |
Low |
LOH |
Undescribed |
Tumor suppressor |
[63] |
|
Bladder |
SALL3 |
Low |
Hypermethylation |
SALL3, CFTR, and TWIST1 are proposed as disease recurrence predictors |
Tumor suppressor |
|
|
Testicular tumors |
SALL4 |
High |
Undescribed |
SALL4 is a novel sensitive and specific marker for testicular germ cell tumors |
Oncogene |
[122] |
|
Kidney |
SALL1 |
Low |
miR-942 |
SALL1 inhibition plays a potential role in sunitinib resistance in RCC patients |
Tumor suppressor |
[108] |
|
Wilms’ tumor |
SALL1 |
High |
Undescribed |
Undescribed |
Oncogene |
|
|
Wilms’ tumor |
SALL2 |
High |
Undescribed |
SALL2 was identified as one of the 27 signature genes highly expressed by comparing tumor samples with normal fetal kidneys |
Oncogene |
[125] |
|
Kidney |
SALL3 |
Low |
Methylation |
SALL3 downregulation may contribute to genome hypermethylation similar to VHL |
Tumor suppressor |
[126] |
|
Wilms’ tumor |
SALL4 |
High |
Undescribed |
Undescribed |
Oncogene |
[127] |
6. Targeting SALLs for Cancer Therapy
References
- De Celis, J.F.; Barrio, R. Regulation and function of Spalt proteins during animal development. Int. J. Dev. Biol. 2009, 53, 1385–1398.
- Sweetman, D.; Munsterberg, A. The vertebrate spalt genes in development and disease. Dev. Biol. 2006, 293, 285–293.
- Hermosilla, V.; Hepp, M.I.; Escobar, D.; Farkas, C.; Riffo, E.N.; Castro, A.F.; Pincheira, R. Developmental SALL2 transcription factor: A new player in cancer. Carcinogenesis 2017, 38, 680–690.
- Lauberth, S.; Rauchman, M. A Conserved 12-Amino Acid Motif in Sall1 Recruits the Nucleosome Remodeling and Deacetylase Corepressor Complex. J. Biol. Chem. 2006, 281, 23922–23931.
- Lorente-Sorolla, J.; Truchado-Garcia, M.; Perry, K.J.; Henry, J.Q.; Grande, C. Molecular, phylogenetic and developmental analyses of Sall proteins in bilaterians. EvoDevo 2018, 9, 9.
- Kawakami, Y.; Uchiyama, Y.; Esteban, C.R.; Inenaga, T.; Koyano-Nakagawa, N.; Kawakami, H.; Marti, M.; Kmita, M.; Monaghan-Nichols, P.; Nishinakamura, R.; et al. Sall genes regulate region-specific morphogenesis in the mouse limb by modulating Hox activities. Development 2009, 136, 585–594.
- Kohlhase, J.; Heinrich, M.; Schubert, L.; Liebers, M.; Kispert, A.; Laccone, F.; Turnpenny, P.; Winter, R.M.; Reardon, W. Okihiro syndrome is caused by SALL4 mutations. Hum. Mol. Genet. 2002, 11, 2979–2987.
- Kelberman, D.; Islam, L.; Lakowski, J.; Bacchelli, C.; Chanudet, E.; Lescai, F.; Patel, A.; Stupka, E.; Buck, A.; Wolf, S.; et al. Mutation of SALL2 causes recessive ocular coloboma in humans and mice. Hum. Mol. Genet. 2014, 23, 2511–2526.
- Dostal, A.; Nemeckova, J.; Gaillyova, R. The 18q deletion syndrome and analysis of the critical region for orofacial cleft at 18q22.3. J. Cranio-Maxillofac. Surg. 2009, 37, 272–275.
- Kiefer, S.M.; Ohlemiller, K.K.; Yang, J.; McDill, B.W.; Kohlhase, J.; Rauchman, M. Expression of a truncated Sall1 transcriptional repressor is responsible for Townes-Brocks syndrome birth defects. Hum. Mol. Genet. 2003, 12, 2221–2227.
- Parrish, M.; Ott, T.; Lance-Jones, C.; Schuetz, G.; Schwaeger-Nickolenko, A.; Monaghan, A.P. Loss of the Sall3 Gene Leads to Palate Deficiency, Abnormalities in Cranial Nerves, and Perinatal Lethality. Mol. Cell. Biol. 2004, 24, 7102–7112.
- Warren, M.; Wang, W.; Spiden, S.; Chen-Murchie, D.; Tannahill, D.; Steel, K.P.; Bradley, A. ASall4 mutant mouse model useful for studying the role ofSall4 in early embryonic development and organogenesis. Genes 2007, 45, 51–58.
- Sato, A.; Matsumoto, Y.; Koide, U.; Kataoka, Y.; Yoshida, N.; Yokota, T.; Asashima, M.; Nishinakamura, R. Zinc Finger Protein Sall2 Is Not Essential for Embryonic and Kidney Development. Mol. Cell. Biol. 2003, 23, 62–69.
- Böhm, J.; Buck, A.; Borozdin, W.; Mannan, A.U.; Matysiak-Scholze, U.; Adham, I.; Schulz-Schaeffer, W.; Floss, T.; Wurst, W.; Kohlhase, J.; et al. Sall1, Sall2, and Sall4 Are Required for Neural Tube Closure in Mice. Am. J. Pathol. 2008, 173, 1455–1463.
- Chai, L. The role of HSAL (SALL) genes in proliferation and differentiation in normal hematopoiesis and leukemogenesis. Transfusion 2011, 51, 87S–93S.
- Hermosilla, V.; Salgado, G.; Riffo, E.; Escobar, D.; Hepp, M.; Farkas, C.; Galindo, M.; Morín, V.; García-Robles, M.A.; Castro, A.F.; et al. SALL2 represses cyclins D1 and E1 expression and restrains G1/S cell cycle transition and cancer-related phenotypes. Mol. Oncol. 2018, 12, 1026–1046.
- Böhm, J.; Sustmann, C.; Wilhelm, C.; Kohlhase, J. SALL4 is directly activated by TCF/LEF in the canonical Wnt signaling pathway. Biochem. Biophys. Res. Commun. 2006, 348, 898–907.
- Basta, J.M.; Singh, A.P.; Robbins, L.; Stout, L.; Pherson, M.; Rauchman, M. The core SWI/SNF catalytic subunit Brg1 regulates nephron progenitor cell proliferation and differentiation. Dev. Biol. 2020, 464, 176–187.
- Basta, J.M.; Robbins, L.; Denner, D.R.; Kolar, G.R.; Rauchman, M. Sall1-NuRD interaction regulates multipotent nephron progenitors and is required for loop of Henle formation. Development 2017, 144, 3080–3094.
- Basta, J.; Rauchman, M. The Nucleosome Remodeling and Deacetylase (NuRD) Complex in Development and Disease. Transl. Res. 2015, 165, 36–47.
- Tahara, N.; Kawakami, H.; Chen, K.Q.; Anderson, A.; Peterson, M.Y.; Gong, W.; Shah, P.; Hayashi, S.; Nishinakamura, R.; Nakagawa, Y.; et al. Sall4 regulates neuromesodermal progenitors and their descendants during body elongation in mouse embryos. Development 2019, 146, 1–10.
- Lai, A.Y.; Wade, P.A. Cancer biology and NuRD: A multifaceted chromatin remodelling complex. Nat. Rev. Cancer 2011, 11, 588–596.
- Lu, J.; Jeong, H.; Kong, N.; Yang, Y.; Carroll, J.; Luo, H.R.; Silberstein, L.E.; Ma, Y.; Chai, L. Stem Cell Factor SALL4 Represses the Transcriptions of PTEN and SALL1 through an Epigenetic Repressor Complex. PLoS ONE 2009, 4, e5577.
- Chan, A.-L.; La, H.M.; Legrand, J.; Mäkelä, J.-A.; Eichenlaub, M.; De Seram, M.; Ramialison, M.; Hobbs, R.M. Germline Stem Cell Activity Is Sustained by SALL4-Dependent Silencing of Distinct Tumor Suppressor Genes. Stem Cell Rep. 2017, 9, 956–971.
- Miller, A.; Ralser, M.; Kloet, S.; Loos, R.; Nishinakamura, R.; Bertone, P.; Vermeulen, M.; Hendrich, B. Sall4 controls differentiation of pluripotent cells independently of the Nucleosome Remodelling and Deacetylation (NuRD) complex. Development 2016, 143, 3074–3084.
- Lauberth, S.M.; Bilyeu, A.C.; Firulli, B.A.; Kroll, K.L.; Rauchman, M. A Phosphomimetic Mutation in the Sall1 Repression Motif Disrupts Recruitment of the Nucleosome Remodeling and Deacetylase Complex and Repression of Gbx2. J. Biol. Chem. 2007, 282, 34858–34868.
- Ma, C.; Wang, F.; Han, B.; Zhong, X.; Si, F.; Ye, J.; Hsueh, E.C.; Robbins, L.; Kiefer, S.M.; Zhang, Y.; et al. SALL1 functions as a tumor suppressor in breast cancer by regulating cancer cell senescence and metastasis through the NuRD complex. Mol. Cancer 2018, 17, 1–21.
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196.
- Li, D.; Tian, Y.; Ma, Y.; Benjamin, T. p150 Sal2 Is a p53-Independent Regulator of p21 WAF1/CIP. Mol. Cell. Biol. 2004, 24, 3885–3893.
- Wu, Z.; Cheng, K.; Shi, L.; Li, Z.; Negi, H.; Gao, G.; Kamle, S.; Li, D. Sal-like protein 2 upregulates p16 expression through a proximal promoter element. Cancer Sci. 2015, 106, 253–261.
- Miao, F.; Zhang, X.; Cao, Y.; Wang, Y.; Zhang, X. Effect of siRNA-silencing of SALL2 gene on growth, migration and invasion of human ovarian carcinoma A2780 cells. BMC Cancer 2017, 17, 838.
- Chen, M.; Li, L.; Zheng, P. SALL4 promotes the tumorigenicity of cervical cancer cells through activation of the Wnt/β-catenin pathway viaCTNNB1. Cancer Sci. 2019, 110, 2794–2805.
- He, J.; Zhou, M.; Chen, X.; Yue, D.; Yang, L.; Qin, G.; Zhang, Z.; Gao, Q.; Wang, D.; Zhang, C.; et al. Inhibition of SALL4 reduces tumorigenicity involving epithelial-mesenchymal transition via Wnt/β-catenin pathway in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2016, 35, 1–13.
- Li, J.; Zhang, Y.; Tao, X.; You, Q.; Tao, Z.; He, Z.; Ou, J. Knockdown of SALL4 inhibits the proliferation, migration, and invasion of human lung cancer cells in vivo and in vitro. Ann. Transl. Med. 2020, 8, 1678.
- Liu, C.; Wu, H.; Li, Y.; Shen, L.; Yu, R.; Yin, H.; Sun, T.; Sun, C.; Zhou, Y.; Du, Z. SALL4 suppresses PTEN expression to promote glioma cell proliferation via PI3K/AKT signaling pathway. J. Neuro-Oncol. 2017, 135, 263–272.
- Liu, C.; Yao, F.; Mao, X.; Li, W.; Chen, H. Effect of SALL4 on the Proliferation, Invasion and Apoptosis of Breast Cancer Cells. Technol. Cancer Res. Treat. 2020, 19, 1–10.
- Sung, C.K.; Yim, H.; Gu, H.; Li, D.; Andrews, E.; Duraisamy, S.; Li, C.; Drapkin, R.; Benjamin, T. The Polyoma Virus Large T Binding Protein p150 Is a Transcriptional Repressor of c-MYC. PLoS ONE 2012, 7, e46486.
- Ma, Y.; Cui, W.; Yang, J.; Qu, J.; Di, C.; Amin, H.M.; Lai, R.; Ritz, J.; Krause, D.S.; Chai, L. SALL4, a novel oncogene, is constitutively expressed in human acute myeloid leukemia (AML) and induces AML in transgenic mice. Blood 2006, 108, 2726–2735.
- Salman, H.; Shuai, X.; Nguyen-Lefebvre, A.T.; Giri, B.; Ren, M.; Rauchman, M.; Robbins, L.; Hou, W.; Korkaya, H.; Ma, Y. SALL1 expression in acute myeloid leukemia. Oncotarget 2018, 9, 7442–7452.
- Sato, A.; Kishida, S.; Tanaka, T.; Kikuchi, A.; Kodama, T.; Asashima, M.; Nishinakamura, R. Sall1, a causative gene for Townes–Brocks syndrome, enhances the canonical Wnt signaling by localizing to heterochromatin. Biochem. Biophys. Res. Commun. 2004, 319, 103–113.
- Yang, J.; Chai, L.; Gao, C.; Fowles, T.C.; Alipio, Z.; Dang, H.; Xu, D.; Fink, L.M.; Ward, D.C.; Ma, Y. SALL4 is a key regulator of survival and apoptosis in human leukemic cells. Blood 2008, 112, 805–813.
- Gu, H.; Li, D.; Sung, C.K.; Yim, H.; Troke, P.; Benjamin, T. DNA-binding and regulatory properties of the transcription factor and putative tumor suppressor p150Sal2. Biochim. Biophys. Acta (BBA)-Bioenerg. 2011, 1809, 276–283.
- Escobar, D.; Hepp, M.; Farkas, C.; Campos, T.M.C.; Sodir, N.M.; Morales, M.M.; Alvarez, C.; Swigart, L.B.; Evan, G.I.; Gutierrez, J.; et al. Sall2 is required for proapoptotic Noxa expression and genotoxic stress-induced apoptosis by doxorubicin. Cell Death Dis. 2015, 6, e1816.
- Hepp, M.I.; Escobar, D.; Farkas, C.; Hermosilla, V.; Álvarez, C.; Amigo, R.; Gutiérrez, J.L.; Castro, A.F.; Pincheira, R. A Trichostatin A (TSA)/Sp1-mediated mechanism for the regulation of SALL2 tumor suppressor in Jurkat T cells. Biochim. Biophys. Acta (BBA)-Bioenerg. 2018, 1861, 623–636.
- Hopkins, B.D.; Hodakoski, C.; Barrows, D.; Mense, S.M.; Parsons, R.E. PTEN function: The long and the short of it. Trends Biochem. Sci. 2014, 39, 183–190.
- Deng, G.; Zhu, L.; Huang, F.; Nie, W.; Huang, W.; Xu, H.; Zheng, S.; Yi, Z.; Wan, T. Knockdown of Sall4 inhibits intrahepatic cholangiocarcinoma cell migration and invasion in ICC-9810 cells. OncoTargets Ther. 2016, 9, 5297–5305.
- Ye, L.; Lin, C.; Wang, X.; Li, Q.; Li, Y.; Wang, M.; Zhao, Z.; Wu, X.; Shi, D.; Xiao, Y.; et al. Epigenetic silencing of SALL 2 confers tamoxifen resistance in breast cancer. EMBO Mol. Med. 2019, 11, e10638.
- Luo, J.; Wang, W.; Tang, Y.; Zhou, D.; Gao, Y.; Zhang, Q.; Zhou, X.; Zhu, H.; Xing, L.; Yu, J. mRNA and methylation profiling of radioresistant esophageal cancer cells: The involvement of Sall2 in acquired aggressive phenotypes. J. Cancer 2017, 8, 646–656.
- Yang, J.; Aguila, J.R.; Alipio, Z.; Lai, R.; Fink, L.M.; Ma, Y. Enhanced self-renewal of hematopoietic stem/progenitor cells mediated by the stem cell gene Sall4. J. Hematol. Oncol. 2011, 4, 38.
- Fukuda, S.; Hoggatt, J.; Singh, P.; Abe, M.; Speth, J.; Hu, P.; Conway, E.; Nucifora, G.; Yamaguchi, S.; Pelus, L.M. Survivin modulates genes with divergent molecular functions and regulates proliferation of hematopoietic stem cells through Evi-1. Leukemia 2014, 29, 433–440.
- Zhou, Q.; Guo, Y.; Zheng, B.; Shao, B.; Jiang, M.; Wang, G.; Zhou, T.; Wang, L.; Zhou, Z.; Guo, X.; et al. Establishment of a proteome profile and identification of molecular markers for mouse spermatogonial stem cells. J. Cell. Mol. Med. 2014, 19, 521–534.
- Morita, Y.; Andersen, P.; Hotta, A.; Tsukahara, Y.; Sasagawa, N.; Hayashida, N.; Koga, C.; Nishikawa, M.; Saga, Y.; Evans, S.M.; et al. Sall1 transiently marks undifferentiated heart precursors and regulates their fate. J. Mol. Cell. Cardiol. 2016, 92, 158–162.
- Osafune, K.; Takasato, M.; Kispert, A.; Asashima, M.; Nishinakamura, R. Identification of multipotent progenitors in the embryonic mouse kidney by a novel colony-forming assay. Development 2006, 133, 151–161.
- Lin, Y.; Xiao, Y.; Lin, C.; Zhang, Q.; Zhang, S.; Pei, F.; Liu, H.; Chen, Z. SALL1 regulates commitment of odontoblast lineages by interacting with RUNX2 to remodel open chromatin regions. Stem Cells 2020, 39, 196–209.
- Hobbs, R.; Fagoonee, S.; Papa, A.; Webster, K.; Altruda, F.; Nishinakamura, R.; Chai, L.; Pandolfi, P.P. Functional Antagonism between Sall4 and Plzf Defines Germline Progenitors. Cell Stem Cell 2012, 10, 284–298.
- Yang, J.; Gao, C.; Chai, L.; Ma, Y. A Novel SALL4/OCT4 Transcriptional Feedback Network for Pluripotency of Embryonic Stem Cells. PLoS ONE 2010, 5, e10766.
- Pantier, R.; Chhatbar, K.; Quante, T.; Skourti-Stathaki, K.; Cholewa-Waclaw, J.; Alston, G.; Alexander-Howden, B.; Lee, H.Y.; Cook, A.G.; Spruijt, C.G.; et al. SALL4 controls cell fate in response to DNA base composition. Mol. Cell 2021, 81, 845–858.e8.
- Suvà, M.L.; Rheinbay, E.; Gillespie, S.M.; Patel, A.P.; Wakimoto, H.; Rabkin, S.D.; Riggi, N.; Chi, A.S.; Cahill, D.P.; Nahed, B.V.; et al. Reconstructing and Reprogramming the Tumor-Propagating Potential of Glioblastoma Stem-like Cells. Cell 2014, 157, 580–594.
- Karantzali, E.; Lekakis, V.; Ioannou, M.; Hadjimichael, C.; Papamatheakis, J.; Kretsovali, A. Sall1 Regulates Embryonic Stem Cell Differentiation in Association with Nanog. J. Biol. Chem. 2011, 286, 1037–1045.
- Quevedo, M.; Meert, L.; Dekker, M.R.; Dekkers, D.H.W.; Brandsma, J.H.; Berg, D.L.C.V.D.; Ozgür, Z.; Van Ijcken, W.F.J.; Demmers, J.; Fornerod, M.; et al. Mediator complex interaction partners organize the transcriptional network that defines neural stem cells. Nat. Commun. 2019, 10, 1–15.
- Argos, M.; Kibriya, M.G.; Jasmine, F.; Olopade, O.I.; Su, T.; Hibshoosh, H.; Ahsan, H. Genomewide scan for loss of heterozygosity and chromosomal amplification in breast carcinoma using single-nucleotide polymorphism arrays. Cancer Genet. Cytogenet. 2008, 182, 69–74.
- Mathew, R.; Arora, S.; Mathur, M.; Ralhan, R.; Chattopadhyay, T.K. Esophageal squamous cell carcinomas with DNA replication errors (RER +) are associated with p16/pRb loss and wild-type p53. J. Cancer Res. Clin. Oncol. 2001, 127, 603–612.
- Chang, W.Y.; Cairns, P.; Schoenberg, M.P.; Polascik, T.J.; Sidransky, D. Novel suppressor loci on chromosome 14q in primary bladder cancer. Cancer Res. 1995, 55, 3246–3249.
- Abujiang, P.; Mori, T.J.; Takahashi, T.; Tanaka, F.; Kasyu, I.; Hitomi, S.; Hiai, H. Loss of heterozygosity (LOH) at 17q and 14q in human lung cancers. Oncogene 1998, 17, 3029–3033.
- Al-Mulla, F.; Alfadhli, S.; Al-Hakim, A.H.; Going, J.J.; Bitar, M.S. Metastatic recurrence of early-stage colorectal cancer is linked to loss of heterozygosity on chromosomes 4 and 14q. J. Clin. Pathol. 2006, 59, 624–630.
- Beder, L.B.; Gunduz, M.; Ouchida, M.; Fukushima, K.; Gunduz, E.; Ito, S.; Sakai, A.; Nagai, N.; Nishizaki, K.; Shimizu, K. Genome-Wide Analyses on Loss of Heterozygosity in Head and Neck Squamous Cell Carcinomas. Lab. Investig. 2003, 83, 99–105.
- Lee, D.J.; Koch, W.M.; Yoo, G.; Lango, M.; Reed, A.; Califano, J.; Brennan, J.A.; Westra, W.H.; Zahurak, M.; Sidransky, D. Impact of chromosome 14q loss on survival in primary head and neck squamous cell carcinoma. Clin. Cancer Res. 1997, 3, 501–505.
- Pehlivan, D.; Gunduz, E.; Gunduz, M.; Nagatsuka, H.; Beder, L.B.; Cengiz, B.; Rivera, R.S.; Fukushima, K.; Palanduz, S.; Ozturk, S.; et al. Loss of heterozygosity at chromosome 14q is associated with poor prognosis in head and neck squamous cell carcinomas. J. Cancer Res. Clin. Oncol. 2008, 134, 1267–1276.
- Nishizuka, S.; Tamura, G.; Terashima, M.; Satodate, R. Loss of heterozygosity during the development and progression of differentiated adenocarcinoma of the stomach. J. Pathol. 1998, 185, 38–43.
- Takebayashi, S.; Hickson, A.; Ogawa, T.; Jung, K.-Y.; Mineta, H.; Ueda, Y.; Grénman, R.; Fisher, S.G.; Carey, T.E. Loss of chromosome arm 18q with tumor progression in head and neck squamous cancer. Genes Chromosomes Cancer 2004, 41, 145–154.
- Jen, J.; Kim, H.; Piantadosi, S.; Liu, Z.-F.; Levitt, R.C.; Sistonen, P.; Kinzler, K.W.; Vogelstein, B.; Hamilton, S.R. Allelic Loss of Chromosome 18q and Prognosis in Colorectal Cancer. N. Engl. J. Med. 1994, 331, 213–221.
- Savelieva, E.; Belair, C.D.; Newton, M.A.; Devries, S.; Gray, J.W.; Waldman, F.; Reznikoff, C.A. 20q gain associates with immortalization: 20q13.2 amplification correlates with genome instability in human papillomavirus 16 E7 transformed human uroepithelial cells. Oncogene 1997, 14, 551–560.
- Werner, M.; Mattis, A.; Aubele, M.; Cummings, M.; Zitzelsberger, H.; Hutzler, P.; Höfler, H. 20q13.2 Amplification in intraductal hyperplasia adjacent to in situ and invasive ductal carcinoma of the breast. Virchows Arch. 1999, 435, 469–472.
- Huang, H.-N.; Huang, W.-C.; Lin, C.-H.; Chiang, Y.-C.; Huang, H.-Y.; Kuo, K.-T. Chromosome 20q13.2 ZNF217 locus amplification correlates with decreased E-cadherin expression in ovarian clear cell carcinoma with PI3K-Akt pathway alterations. Hum. Pathol. 2014, 45, 2318–2325.
- Briand-Suleau, A.; Martinovic, J.; Tosca, L.; Tou, B.; Brisset, S.; Bouligand, J.; Delattre, V.; Giurgea, I.; Bachir, J.; Folliot, P.; et al. SALL4 and NFATC2: Two major actors of interstitial 20q13.2 duplication. Eur. J. Med. Genet. 2014, 57, 174–180.
- Okamoto, A.; Sehouli, J.; Yanaihara, N.; Hirata, Y.; Braicu, I.; Kim, B.-G.; Takakura, S.; Saito, M.; Yanagida, S.; Takenaka, M.; et al. Somatic Copy Number Alterations Associated with Japanese or Endometriosis in Ovarian Clear Cell Adenocarcinoma. PLoS ONE 2015, 10, e0116977.
- Koh, H.M.; Jang, B.G.; Hyun, C.L.; Kim, Y.S.; Hyun, J.W.; Chang, W.Y.; Maeng, Y.H. Aurora Kinase A Is a Prognostic Marker in Colorectal Adenocarcinoma. J. Pathol. Transl. Med. 2017, 51, 32–39.
- Morikawa, A.; Hayashi, T.; Kobayashi, M.; Kato, Y.; Shirahige, K.; Itoh, T.; Urashima, M.; Okamoto, A.; Akiyama, T. Somatic copy number alterations have prognostic impact in patients with ovarian clear cell carcinoma. Oncol. Rep. 2018, 40, 309–318.
- Sung, C.K.; Dahl, J.; Yim, H.; Rodig, S.; Benjamin, A.T.L. Transcriptional and post-translational regulation of the quiescence factor and putative tumor suppressor p150Sal2. FASEB J. 2011, 25, 1275–1283.
- Ma, Y.; Li, D.; Chai, L.; Luciani, A.M.; Ford, D.; Morgan, J.; Maizel, A.L. Cloning and Characterization of Two Promoters for the Human HSAL2 Gene and Their Transcriptional Repression by the Wilms Tumor Suppressor Gene Product. J. Biol. Chem. 2001, 276, 48223–48230.
- Farkas, C.; Martins, C.P.; Escobar, D.; Hepp, M.I.; Donner, D.B.; Castro, A.F.; Evan, G.; Gutiérrez, J.L.; Warren, R.; Pincheira, R. Wild Type p53 Transcriptionally Represses the SALL2 Transcription Factor under Genotoxic Stress. PLoS ONE 2013, 8, e73817.
- Pecce, V.; Verrienti, A.; Fiscon, G.; Sponziello, M.; Conte, F.; Abballe, L.; Durante, C.; Farina, L.; Filetti, S.; Paci, P. The role of FOSL1 in stem-like cell reprogramming processes. Sci. Rep. 2021, 11, 1–11.
- Bard, J.D.; Gelebart, P.; Amin, H.M.; Young, L.C.; Ma, Y.; Lai, R. Signal transducer and activator of transcription 3 is a transcriptional factor regulating the gene expression ofSALL4. FASEB J. 2009, 23, 1405–1414.
- Fujii, Y.; Yoshihashi, K.; Suzuki, H.; Tsutsumi, S.; Mutoh, H.; Maeda, S.; Yamagata, Y.; Seto, Y.; Aburatani, H.; Hatakeyama, M. CDX1 confers intestinal phenotype on gastric epithelial cells via induction of stemness-associated reprogramming factors SALL4 and KLF5. Proc. Natl. Acad. Sci. USA 2012, 109, 20584–20589.
- Hill, V.K.; Hesson, L.B.; Dansranjavin, T.; Dallol, A.; Bieche, I.; Vacher, S.; Tommasi, S.; Dobbins, T.; Gentle, D.; Euhus, D.; et al. Identification of 5 novel genes methylated in breast and other epithelial cancers. Mol. Cancer 2010, 9, 51.
- Wang, C.; Pu, W.; Zhao, D.; Zhou, Y.; Lu, T.; Chen, S.; He, Z.; Feng, X.; Wang, Y.; Li, C.; et al. Identification of Hyper-Methylated Tumor Suppressor Genes-Based Diagnostic Panel for Esophageal Squamous Cell Carcinoma (ESCC) in a Chinese Han Population. Front. Genet. 2018, 9, 356.
- Imai, A.; Mochizuki, D.; Misawa, Y.; Nakagawa, T.; Endo, S.; Mima, M.; Yamada, S.; Kawasaki, H.; Kanazawa, T.; Misawa, K. SALL2 Is a Novel Prognostic Methylation Marker in Patients with Oral Squamous Carcinomas: Associations with SALL1 and SALL3 Methylation Status. DNA Cell Biol. 2019, 38, 678–687.
- Misawa, K.; Kanazawa, T.; Mochizuki, D.; Imai, A.; Mima, M.; Yamada, S.; Morita, K.; Misawa, Y.; Shinmura, K.; Mineta, H. Genes Located on 18q23 Are Epigenetic Markers and Have Prognostic Significance for Patients with Head and Neck Cancer. Cancers 2019, 11, 401.
- Misawa, K.; Mochizuki, D.; Imai, A.; Misawa, Y.; Endo, S.; Mima, M.; Kawasaki, H.; Carey, T.E.; Kanazawa, T. Epigenetic silencing of SALL3 is an independent predictor of poor survival in head and neck cancer. Clin. Epigenet. 2017, 9, 1–12.
- Misawa, K.; Misawa, Y.; Imai, A.; Mochizuki, D.; Endo, S.; Mima, M.; Ishikawa, R.; Kawasaki, H.; Yamatodani, T.; Kanazawa, T. Epigenetic modification of SALL1 as a novel biomarker for the prognosis of early stage head and neck cancer. J. Cancer 2018, 9, 941–949.
- Lin, J.; Qian, J.; Yao, D.-M.; Qian, W.; Yang, J.; Wang, C.-Z.; Chai, H.-Y.; Ma, J.-C.; Deng, Z.-Q.; Li, Y.; et al. Aberrant hypomethylation of SALL4 gene in patients with myelodysplastic syndrome. Leuk. Res. 2013, 37, 71–75.
- Ma, J.-C.; Qian, J.; Lin, J.; Qian, W.; Yang, J.; Wang, C.-Z.; Chai, H.-Y.; Li, Y.; Chen, Q.; Qian, Z. Aberrant hypomethylation of SALL4 gene is associated with intermediate and poor karyotypes in acute myeloid leukemia. Clin. Biochem. 2013, 46, 304–307.
- Liu, J.; Sauer, M.A.; Hussein, S.G.; Yang, J.; Tenen, D.G.; Chai, L. SALL4 and microRNA: The Role of Let-7. Genes 2021, 12, 1301.
- Yang, C.M.; Chiba, T.; Brill, B.; Delis, N.; von Manstein, V.; Vafaizadeh, V.; Oellerich, T.; Groner, B. Expression of the mi R-302/367 cluster in glioblastoma cells suppresses tumorigenic gene expression patterns and abolishes transformation related phenotypes. Int. J. Cancer 2015, 137, 2296–2309.
- Zhou, Y.; Liu, Y.; Hu, C.; Jiang, Y. MicroRNA-16 inhibits the proliferation, migration and invasion of glioma cells by targeting Sal-like protein 4. Int. J. Mol. Med. 2016, 38, 1768–1776.
- Chen, L.-P.; Zhang, N.-N.; Ren, X.-Q.; He, J.; Li, Y. miR-103/miR-195/miR-15b Regulate SALL4 and Inhibit Proliferation and Migration in Glioma. Molecules 2018, 23, 2938.
- He, J.; Zhang, W.; Zhou, Q.; Zhao, T.; Song, Y.; Chai, L.; Li, Y. Low-expression of microRNA-107 inhibits cell apoptosis in glioma by upregulation of SALL4. Int. J. Biochem. Cell Biol. 2013, 45, 1962–1973.
- Zhou, Y.; Peng, Y.; Liu, M.; Jiang, Y. MicroRNA-181b Inhibits Cellular Proliferation and Invasion of Glioma Cells via Targeting Sal-Like Protein 4. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2017, 25, 947–957.
- Wang, M.; Qiu, R.; Gong, Z.; Zhao, X.; Wang, T.; Zhou, L.; Lu, W.; Shen, B.; Zhu, W.; Xu, W. miR-188-5p emerges as an oncomiRNA to promote gastric cancer cell proliferation and migration via upregulation of SALL4. J. Cell. Biochem. 2019, 120, 15027–15037.
- Jiang, X.; Wang, Z. miR-16 targets SALL4 to repress the proliferation and migration of gastric cancer. Oncol. Lett. 2018, 16, 3005–3012.
- Kondelova, A.; Alburquerque-González, B.; Vychytilova-Faltejskova, P.; García-Solano, J.; Prochazka, V.; Kala, Z.; Pérez, F.; Slaby, O.; Conesa-Zamora, P. miR-181a-2* expression is different amongst carcinomas from the colorectal serrated route. Mutagenesis 2020, 35, 233–241.
- Cheng, J.; Deng, R.; Zhang, P.; Wu, C.; Wu, K.; Shi, L.; Liu, X.; Bai, J.; Deng, M.; Shuai, X.; et al. miR-219-5p plays a tumor suppressive role in colon cancer by targeting oncogene Sall4. Oncol. Rep. 2015, 34, 1923–1932.
- Chang, S.; Sun, G.; Zhang, D.; Li, Q.; Qian, H. MiR-3622a-3p acts as a tumor suppressor in colorectal cancer by reducing stemness features and EMT through targeting spalt-like transcription factor 4. Cell Death Dis. 2020, 11, 1–19.
- Rahnama, M.A.; Movassaghpour, A.A.; Soleimani, M.; Atashi, A.; Anbarlou, A.; Asenjan, K.S. MicroRNA-15b target Sall4 and diminish in vitro UCB-derived HSCs expansion. EXCLI J. 2015, 14, 601–610.
- Melton, C.; Judson, R.L.; Blelloch, R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nat. Cell Biol. 2010, 463, 621–626.
- Liu, X.; Cao, Y.; Zhang, Y.; Zhou, H.; Li, H. Regulatory effect of MiR103 on proliferation, EMT and invasion of oral squamous carcinoma cell through SALL4. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9931–9938.
- Yuan, J.H.; Li, W.X.; Hu, C.; Zhang, B. Upregulation of SNHG12 accelerates cell proliferation, migration, invasion and restrain cell apoptosis in breast cancer by enhancing regulating SALL4 expression via sponging miR-15a-5p. Neoplasma 2020, 67, 861–870.
- Chen, Y.; He, J.; Su, C.; Wang, H.; Chen, Y.; Guo, W.; Li, Y.; Ding, G. LINC00461 affects the survival of patients with renal cell carcinoma by acting as a competing endogenous RNA for microRNA-942. Oncol. Rep. 2019, 42, 1924–1934.
- Li, Z.; Zhao, S.; Wang, H.; Zhang, B.; Zhang, P. miR-4286 promotes prostate cancer progression via targeting the expression of SALL1. J. Gene Med. 2019, e3127.
- Xia, H.; Niu, Q.; Ding, Y.; Zhang, Z.; Yuan, J.; Jin, W. Long noncoding HOXA11-AS knockdown suppresses the progression of non-small cell lung cancer by regulating miR-3619-5p/SALL4 axis. J. Mol. Histol. 2021, 52, 729–740.
- Wang, Y.; Liu, N.; Li, M.-Y.; Du, M.-F. Long non-coding RNA ZEB2-AS1 regulates osteosarcoma progression by acting as a molecular sponge of miR-107 to modulate SALL4 expression. Am. J. Transl. Res. 2021, 13, 1140–1154.
- Shi, D.-M.; Shi, X.-L.; Xing, K.-L.; Zhou, H.-X.; Lu, L.-L.; Wu, W.-Z. miR-296-5p suppresses stem cell potency of hepatocellular carcinoma cells via regulating Brg1/Sall4 axis. Cell. Signal. 2020, 72, 109650.
- Ma, Y.-S.; Liu, J.-B.; Lin, L.; Zhang, H.; Wu, J.-J.; Shi, Y.; Jia, C.-Y.; Zhang, D.-D.; Yu, F.; Wang, H.-M.; et al. Exosomal microRNA-15a from mesenchymal stem cells impedes hepatocellular carcinoma progression via downregulation of SALL4. Cell Death Discov. 2021, 7, 1–11.
- Misawa, K.; Misawa, Y.; Mima, M.; Yamada, S.; Imai, A.; Mochizuki, D.; Nakagawa, T.; Kurokawa, T.; Endo, S.; Kawasaki, H.; et al. Overexpression of Sal-like protein 4 in head and neck cancer: Epigenetic effects and clinical correlations. Cell. Oncol. 2020, 43, 631–641.
- Zhang, D.-L.; Qu, L.-W.; Ma, L.; Zhou, Y.-C.; Wang, G.-Z.; Zhao, X.-C.; Zhang, C.; Zhang, Y.-F.; Wang, M.; Zhang, M.-Y.; et al. Genome-wide identification of transcription factors that are critical to non-small cell lung cancer. Cancer Lett. 2018, 434, 132–143.
- Gautam, A.K.; Wang, C.; Zeng, J.; Wang, J.; Lu, J.; Wei, J.; Huang, G.; Mo, B.; Luo, M.; Mo, B. Expression and clinical significance of SALL4 and LGR5 in patients with lung cancer. Oncol. Lett. 2015, 10, 3629–3634.
- Yanagihara, N.; Kobayashi, D.; Kuribayashi, K.; Tanaka, M.; Hasegawa, T.; Watanabe, N. Significance of SALL4 as a drug-resistant factor in lung cancer. Int. J. Oncol. 2015, 46, 1527–1534.
- Abnet, C.C.; Arnold, M.; Wei, W.-Q. Epidemiology of Esophageal Squamous Cell Carcinoma. Gastroenterology 2018, 154, 360–373.
- Forghanifard, M.M.; Khales, S.A.; Javdani-Mallak, A.; Rad, A.; Farshchian, M.; Abbaszadegan, M.R. Stemness state regulators SALL4 and SOX2 are involved in progression and invasiveness of esophageal squamous cell carcinoma. Med. Oncol. 2014, 31, 1–8.
- Yu, J.; Zhu, T.; Wang, Z.; Zhang, H.; Qian, Z.; Xu, H.; Gao, B.; Wang, W.; Gu, L.; Meng, J.; et al. A Novel Set of DNA Methylation Markers in Urine Sediments for Sensitive/Specific Detection of Bladder Cancer. Clin. Cancer Res. 2007, 13, 7296–7304.
- Van Der Heijden, A.G.; Mengual, L.; Ingelmo, M.; Lozano, J.J.; Westerlo, C.C.M.V.R.-V.D.; Baixauli, M.; Geavlete, B.; Moldoveanud, C.; Ene, C.; Dinney, C.P.; et al. Urine cell-based DNA methylation classifier for monitoring bladder cancer. Clin. Epigenet. 2018, 10, 71.
- Cao, D.; Humphrey, P.A.; Allan, R.W. SALL4 is a novel sensitive and specific marker for metastatic germ cell tumors, with particular utility in detection of metastatic yolk sac tumors. Cancer 2009, 115, 2640–2651.
- Ma, Y.; Singer, D.B.; Gozman, A.; Ford, D.; Chai, L.; Steinhoff, M.M.; Hansen, K.; Maizel, A.L. Hsal 1 is related to kidney and gonad development and is expressed in Wilms tumor. Pediatr. Nephrol. 2001, 16, 701–709.
- Brown, K.W.; Charles, A.; Dallosso, A.; White, G.; Charlet, J.; Standen, G.R.; Malik, K. Characterization of 17.94, a novel anaplastic Wilms’ tumor cell line. Cancer Genet. 2012, 205, 319–326.
- Li, C.-M.; Guo, M.; Borczuk, A.; Powell, C.A.; Wei, M.; Thaker, H.M.; Friedman, R.; Klein, U.; Tycko, B. Gene Expression in Wilms’ Tumor Mimics the Earliest Committed Stage in the Metanephric Mesenchymal-Epithelial Transition. Am. J. Pathol. 2002, 160, 2181–2190.
- Artemov, A.V.; Zhigalova, N.; Zhenilo, S.; Mazur, A.M.; Prokhortchouk, E.B. VHL inactivation without hypoxia is sufficient to achieve genome hypermethylation. Sci. Rep. 2018, 8, 10667.
- Deisch, J.; Raisanen, J.; Rakheja, D. Immunoexpression of SALL4 in Wilms Tumors and Developing Kidney. Pathol. Oncol. Res. 2011, 17, 639–644.
- Yong, K.J.; Li, A.; Ou, W.-B.; Hong, C.K.Y.; Zhao, W.; Wang, F.; Tatetsu, H.; Yan, B.; Qi, L.; Fletcher, J.A.; et al. Targeting SALL4 by entinostat in lung cancer. Oncotarget 2016, 7, 75425–75440.
- Gao, C.; Dimitrov, T.; Yong, K.J.; Tatetsu, H.; Jeong, H.-W.; Luo, H.R.; Bradner, J.E.; Tenen, D.; Chai, L. Targeting transcription factor SALL4 in acute myeloid leukemia by interrupting its interaction with an epigenetic complex. Blood 2013, 121, 1413–1421.
- Yong, K.J.; Gao, C.; Lim, J.S.; Yan, B.; Yang, H.; Dimitrov, T.; Kawasaki, A.; Ong, C.W.; Wong, K.-F.; Lee, S.; et al. Oncofetal Gene SALL4 in Aggressive Hepatocellular Carcinoma. N. Engl. J. Med. 2013, 368, 2266–2276.
- Liu, B.H.; Jobichen, C.; Chia, C.S.B.; Chan, T.H.M.; Tang, J.P.; Chung, T.X.Y.; Li, J.; Poulsen, A.; Hung, A.W.; Koh-Stenta, C.X.; et al. Targeting cancer addiction for SALL4 by shifting its transcriptome with a pharmacologic peptide. Proc. Natl. Acad. Sci. USA 2018, 115, E7119–E7128.
- Sievers, Q.L.; Petzold, G.; Bunker, R.D.; Renneville, A.; Słabicki, M.; Liddicoat, B.J.; Abdulrahman, W.; Mikkelsen, T.; Ebert, B.L.; Thomä, N.H. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science 2018, 362, eaat0572.
- Donovan, K.A.; An, J.; Nowak, R.P.; Yuan, J.C.; Fink, E.C.; Berry, B.C.; Ebert, B.L.; Fischer, E.S. Thalidomide promotes degradation of SALL4, a transcription factor implicated in Duane Radial Ray syndrome. eLife 2018, 7, 1–25.
- Akiyama, R.; Kawakami, H.; Wong, J.; Oishi, I.; Nishinakamura, R.; Kawakami, Y. Sall4-Gli3 system in early limb progenitors is essential for the development of limb skeletal elements. Proc. Natl. Acad. Sci. USA 2015, 112, 5075–5080.
- Chen, K.Q.; Tahara, N.; Anderson, A.; Kawakami, H.; Kawakami, S.; Nishinakamura, R.; Pandolfi, P.P.; Kawakami, Y. Development of the Proximal-Anterior Skeletal Elements in the Mouse Hindlimb Is Regulated by a Transcriptional and Signaling Network Controlled by Sall4. Genetics 2020, 215, 129–141.




