Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Oscar Molina | + 1830 word(s) | 1830 | 2021-12-28 07:01:47 | | | |
| 2 | Yvaine Wei | + 207 word(s) | 2037 | 2022-01-10 03:11:20 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Molina, O. Near-Haploidy and Low-Hypodiploidy in B-Cell Acute Lymphoblastic Leukemia. Encyclopedia. Available online: https://encyclopedia.pub/entry/17904 (accessed on 08 February 2026).
Molina O. Near-Haploidy and Low-Hypodiploidy in B-Cell Acute Lymphoblastic Leukemia. Encyclopedia. Available at: https://encyclopedia.pub/entry/17904. Accessed February 08, 2026.
Molina, Oscar. "Near-Haploidy and Low-Hypodiploidy in B-Cell Acute Lymphoblastic Leukemia" Encyclopedia, https://encyclopedia.pub/entry/17904 (accessed February 08, 2026).
Molina, O. (2022, January 08). Near-Haploidy and Low-Hypodiploidy in B-Cell Acute Lymphoblastic Leukemia. In Encyclopedia. https://encyclopedia.pub/entry/17904
Molina, Oscar. "Near-Haploidy and Low-Hypodiploidy in B-Cell Acute Lymphoblastic Leukemia." Encyclopedia. Web. 08 January, 2022.
Copy Citation
B-cell acute lymphoblastic leukemia (B-ALL) is characterized by an uncontrolled proliferation of blood cells in the bone marrow. Hypodiploidy with less than 40 chromosomes is a rare genetic abnormality in B-cell acute lymphoblastic leukemia (B-ALL). This condition can be classified based on modal chromosome number as low-hypodiploidy (30–39 chromosomes) and near-haploidy (24–29 chromosomes), with unique cytogenetic and mutational landscapes.
hypodiploidy
near-haploidy
B-cell acute lymphoblastic leukemia
1. Introduction
Acute lymphoblastic leukemia (ALL) is a neoplasm arising from lymphoid precursor cells and can be classified as B-ALL or T-ALL based on the immunophenotype of the neoplastic cells [1]. The global incidence of ALL is ~3 cases per 100,000 people and shows a bimodal distribution, with a predominant peak early in life (1 to 15 years) and a second, much lower, peak in older groups (>55 years) [2] (Figure 1). ALL has a slightly higher incidence in males, with a male-to-female ratio of 1.2:1 [3]. The disease is characterized by the uncontrolled proliferation of leukemic cells, which invade the bone marrow (BM), peripheral blood (PB), and other hematopoietic tissues including spleen, liver, and lymph nodes, resulting in a hematopoietic displacement which is responsible for the cytopenias frequently observed at diagnosis. ALL cells also infiltrate commonly the central nervous system (CNS).
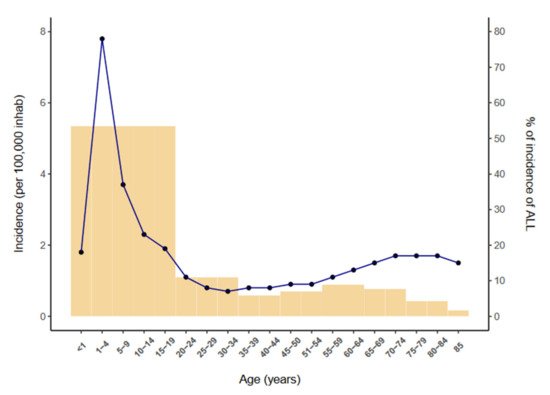
Figure 1. Incidence of ALL per 100,000 inhabitants by age (2014–2018) according to the SEER database [2].
B-cell precursor ALL (B-ALL) accounts for 80–85% of ALL cases and is characterized by small-medium sized leukemic blast cells staining almost always positive for the B-cell antigens CD19, cytoplasmic CD79a and CD22. Although BM and PB are involved in most cases, B-ALL occasionally presents with primary nodal or extranodal sites (B-lymphoblastic lymphoma), which predominantly affect skin, soft tissue, bone and lymph nodes [4].
2. Definition of Hypodiploid B-ALL Subgroups
Hypodiploidy -the loss of one or more whole chromosomes- is a rare cytogenetic finding (≤7%) in children and adults with B-ALL and is generally an adverse prognostic marker [5][6][7][8][9][10][11][12][13][14][15][16]. Most cases (~80%) of hypodiploid B-ALL present with 45 chromosomes and are classified as near-diploid B-ALL, a clinically distinct entity characterized by rearrangements that form dicentric chromosomes but that does not have outcomes as poor as those associated with hypodiploid B-ALL [11].
3. Cytogenetic Characterization of B-ALL with Hypodiploidy <40 Chromosomes
Hypodiploid cases with <40 chromosomes can be further subdivided into two groups based on the bimodal distribution of chromosome numbers: (i) near-haploidy, with 24–29 chromosomes (Figure 2A), and (ii) low-hypodiploidy, with 30–39 chromosomes (Figure 2B,C) [17] (Table 1). Although the modal number of chromosomes is variable, the most recurrent modal numbers are 25–28 for near-haploid and 33–39 for low-hypodiploid B-ALL [18]. Based on conventional chromosome banding analyses, hypodiploidy with <40 chromosomes shows a non-random loss of chromosomes: in near-haploid B-ALL, retained disomies generally comprise chromosomes 8, 10, 14, 18, 21, X and Y [11][12][19][20] whereas in low-hypodiploid cases, retained disomies are more variable and typically comprise chromosomes 1, 5, 6, 8, 10, 11, 14, 18, 19, 21, 22, X and Y, with retained disomies for chromosomes 1, 6, 11 and 18 being the most frequently observed. The most typically lost chromosomes are chromosome 3, 7, 9, 15, 16 and 17 [11][12][19][21][22]. The non-random retention of chromosomes suggests that these chromosomes may harbor specific genes that enhance the oncogenic potential of leukemic cells.
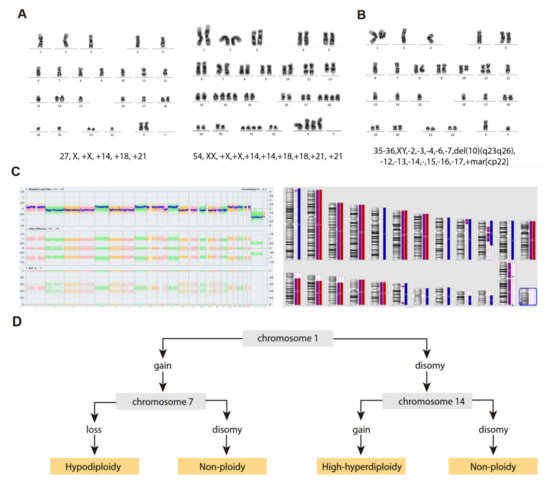
Figure 2. Cytogenetic characterization of B-ALL with <40 chromosomes. (A) G-banded karyotype of near-haploid B-ALL leukemic cells. Left panel, near-haploid clone. Right panel, chromosomally-doubled clone of the same patient. (B) G-banded karyotype of low-hypodiploid B-ALL leukemic cells. Karyotype formulas are indicated below. (C) SNP-array karyogram obtained for the low-hypodiploid B-ALL patient in B. Right panel, blue bars indicate chromosomal disomies of the duplicated/near-triploid clone, red bars indicate chromosomal losses, and purple bars indicate absence of heterozygosity. Left panel, Log2 ratio plot detailing whole chromosomal view for each chromosome, the figure demonstrates pattern of low-hypodiploidy where chromosomes with the lowest Log2 ratio represent the monosomies and a partial deletion of chromosome 10. Allele difference plot and B-allele frequency plot (BAF; BB, AB and AA alleles) indicates copy-neutral loss of heterozygosity. (D) Algorithm proposed by Creasey et al. [22] to distinguish hypodiploid with <40 chromosomes and high-hyperdiploid B-ALL cases based on specific chromosomal gains.
Table 1. Cytogenetic characteristics of B-ALL patients with <40 chromosomes.
| Age (Years) | Near-Haploid | Low-Hypodiploid | Reference | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MN | Retained chr | Lost chr | Doubled Clone | Frequency | MN | Retained chr | Lost chr | Doubled Clone | Frequency | ||
| 1–18 | 25–28 | 8, 10, 14, 18, 21 and sex chr. | yes | 0.0046 | 30–40 | 1, 19, 21, 22 and sex chr. | 3, 7, 13, 16, 17 | yes | 0.41 | [7] | |
| 1–10 | 24–28 | 8, 10, 14, 18, 21 and sex chr. | 7, 13, 14, 20, X | yes | 0.0042 | 33–44 | 7, 13, 14, 20, X | nr | 0.79 | [9] | |
| 2–15 | 23–29 | 14, 18, 21 and sex chr. | yes | 0.0039 | 33–39 | 1, 2, 5, 6, 8, 10, 11,12, 14, 18, 19, 21, 22 and the sex chr. | 7, 17 | yes | 0.39 | [11] | |
| 15–84 | - | - | - | - | - | 30–39 | 1, 5, 6, 8, 10, 11, 15, 18, 19, 21, 22, X, Y | 3, 7, 15, 16, 17 | 66 to 78 chr | 0.05 | [23] |
| 15–55 | - | - | - | - | - | 33–39 | nr | nr | nr | 0.0008 | [11] |
| 15–55/>55 | <30 | 0.0016 | 32–39 | 1 | 2, 3, 7, 9, 13, 15, 16, 17, 20, 4 | 64–74 | 3.85% | [24] | |||
| 1–9/>10 | 24–29 | 14, 18, 21 and sex chr. | nr | nr | nr | 33–39 | nrec | 3, 7, 16, 17 | nr | nr | [25] |
| <31 | 24–31 | 14, 18, 21 and sex chr. | nr | yes | 0.008 | 32–39 | 1, 8, 10, 11, 18, 19, 21 and 22 | nr | nr | 0.0064 | [26] |
Abbreviations: chr, chromosomes; MN, modal numbers; nr, non-reported; nrec, non-recurrent.
“Masked Hypodiploidy”: A Clinical Challenge
A feature common to patients with near-haploid and low-hypodiploid B-ALL is the presence of a clone with an exact or near-exact chromosome doubling of the hypodiploid clone, resulting in a clone with a modal chromosome number of 50–78, in the high-hyperdiploid or triploid range [7][11][12][26] (Table 1) (Figure 2A). Notably, some chromosomes are preferentially lost after chromosome doubling; typically, chromosomes 2, 5, 6, 10, 14 and 22 [21]. The presence of hypodiploid doubled clones has been observed in ~60–65% of patients with near-haploid and low-hypodiploid B-ALL in different studies, and is commonly observed as a mosaic with both hypodiploid and hyperdiploid (doubled) clones visible by standard cytogenetics, fluorescence in situ hybridization (FISH) or flow cytometry analysis of DNA content [11][26][27]. Furthermore, the doubled clone may be the only one detected at diagnosis, leading to the manifestation known as “masked hypodiploidy”, which is clinically challenging since patients can be erroneously classified and treated for high-hyperdiploid B-ALL while being at higher risk of treatment failure. It has been reported that there is no difference in clinical outcome for patients with “masked hypodiploidy”, those who are mosaic for a doubled clone and a hypodiploid clone, and those who have only a hypodiploid clone [12][28][26]. In addition, the hypodiploid clone tends to be quantitatively more frequent at relapse, suggesting that the actual hypodiploid clones may be more chemoresistant than their hyperdiploid (doubled) counterparts [10][29].
4. Molecular Characterization of Hypodiploid B-ALL with <40 Chromosomes
In addition to the massive genetic losses, both near-haploid and low-hypodiploid B-ALL show characteristic and differentiated gene expression profiles, in addition to specific mutational and focal copy-number alteration (CNA) landscapes, those excluding whole chromosome losses [19]. Notably, near-haploid and low-hypodiploid B-ALL presenting with or without doubled clones show similar transcriptional and mutational profiles [19], most likely explaining the similar clinical outcomes between patients with and without chromosomal doubling [7][12][26].
4.1. Near-Haploid B-ALL
The mutational landscape of near-haploid B-ALL is characterized mainly by the presence of alterations involving receptor tyrosine kinases and activating RAS signaling alterations, with >70% of patients showing mutations or focal CNA involving genes in these pathways (Table 2) [19][21][30]. The different RAS signaling alterations have been shown to be mutually exclusive, suggesting that, in contrast to the convergent evolution for RAS mutations observed in infants with MLL-rearranged B-ALL [31], a single alteration in the pathway is sufficient to maintain constitutive RAS-pathway activation. Focal deletions or point mutations in NF1 gene are the most recurrent genetic alterations of near-haploid B-ALL (≥44% of patients) [13][19][32].
Table 2. Molecular characteristics of hypodiploid <40 chromosomes B-ALL (Adapted from Holmfelt et al., 2013 [19]).
| Genes | Cellular Pathway | Near-Haploid B-ALL | Low-Hypodiploid B-ALL | ||||
|---|---|---|---|---|---|---|---|
| Mutation | Focal Deletion | Focal DEL + Mut | Mutation | Focal Deletion | Focal DEL + Mut | ||
| NF1 | RTK/RAS pathway | 11/68 (16%) | 16/68 (24%) | 3/68 (4%) | 0 | 2/34 (6%) | 0 |
| KRAS | 2/68 (3%) | 0 | 0 | 0 | 0 | 0 | |
| NRAS | 10/68 (15%) | 0 | 0 | 0 | 0 | 0 | |
| PTPN11 | 1/68 (1%) | 0 | 0 | 0 | 0 | 0 | |
| FLT3 | 6/68 (9%) | 0 | 0 | 0 | 0 | 0 | |
| CRLF2 | 0 | 2/68 (3%) * | 0 | 0 | 0 | 0 | |
| MAPK1 | 1/68 (1%) | 0 | 0 | 0 | 0 | 0 | |
| GAB2 | 0 | 2/68 (3%) | 0 | 0 | 1/34 (3%) | 0 | |
| EPHA7 | 0 | 2/68 (3%) | 0 | 0 | 0 | 0 | |
| RASA2 | 0 | 2/68 (3%) | 0 | 0 | 0 | 0 | |
| IKZF1 | B-cell development | 0 | 3/68 (4%) | 0 | 0 | 1/34 (3%) | 0 |
| IKZF2 | 1/68 (1%) | 0 | 0 | 0 | 18/34 (53%) | 0 | |
| IKZF3 | 1/68 (1%) | 8/68 (12%) | 0 | 0 | 1/34 (3%) | 0 | |
| PAX5 | 1/68 (1%) | 4/68 (6%) | 0 | 0 | 2/34 (6%) | 0 | |
| EBF1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| VPREB1 | 0 | 3/68 (4%) | 0 | 0 | 2/34 (6%) | 0 | |
| CDKN2A/B | Cell cycle and apoptosis | 0 | 15/68 (22%) | 0 | 0 | 8/34 (24%) | 0 |
| TP53 | 2/68 (3%) | 0 | 0 | 31/34 (91%) | 0 | 0 | |
| RB1 | 2/68 (3%) | 3/68 (4%) | 1/68 (1%) | 5/34 (15%) | 8/34 (24%) | 0 | |
| ETV6 | Hematopoiesis | 1/68 (1%) | 3/68 (4%) | 1/68 (1%) | 0 | 0 | 0 |
| Histone cluster (6p22) | Histone-related | 0 | 13/68 (19%) | 0 | 0 | 1/34 (3%) | 0 |
| ARID1B | 0 | 2/68 (3%) | 0 | 0 | 0 | 0 | |
| PAG1 | BCR signalling | 1/68 (1%) | 6/68 (9%) | 0 | 0 | 1/34 (3%) | 0 |
| ARPP21 | Calmodulin signalling | 0 | 1/68 (1%) | 0 | 0 | 0 | 0 |
| SLX4IP (C20orf194) | Telomere length maintenance | 0 | 2/68 (3%) | 0 | 0 | 0 | 0 |
| CUL5 | Ubiquitin pathway | 0 | 2/68 (3%) | 0 | 0 | 0 | 0 |
| FAM53B | Wnt signalling | 0 | 2/68 (3%) | 0 | 0 | 0 | 0 |
| PDS5B (APRIN) | Cohesin complex | 0 | 2/68 (3%) | 0 | 0 | 0 | 0 |
| ANKRD11 | Cell adhesion | 0 | 0 | 0 | 0 | 2/34 (6%) | 0 |
| DMD | 0 | 0 | 0 | 0 | 1/34 (3%) | 0 | |
* one patient encoding P2RY8-CRLF2.
4.2. Low-Hypodiploid B-ALL
The genetic hallmark of low-hypodiploid B-ALL is TP53 mutations, which are observed in >90% of patients in both childhood and adult low-hypodiploid B-ALL [19][21][30][33]. Most are missense mutations in exons 5–8, affecting the DNA-binding domain and the nuclear localization sequence [19][21]. Other characteristic and recurrent genetic alterations in the low-hypodiploid B-ALL subtype are RB1 mutations or deletions (41% of cases), deletions of IKZF2/Helios (53% of cases) and deletions of CDKN2A/B genes (22% of cases) [19][21]. Mutations of TP53 are found in homozygosity in virtually all low-hypodiploid B-ALL cases due to the very recurrent loss of chromosome 17. TP53 mutations are frequently found in non-tumor hematopoietic cells in 50% of the cases of childhood low-hypodiploid B-ALL [21][30], suggesting that these cases may be a manifestation of Li-Fraumeni syndrome or other germline TP53 cancer-predisposing mutations [19][33][34]. Accordingly, genetic counseling is recommended for children with low-hypodiploid B-ALL carrying TP53 mutations, and their relatives [35][36]. In contrast to childhood cases, TP53 mutations in low-hypodiploid adult B-ALL are somatic, are not found in healthy hematopoietic cells, and not detectable in remission samples [19][21].
5. Etiology of Hypodiploidy in B-ALL
Genomic analyses of these subtypes have been difficult given the limited number of cases; however, a study on a small cohort of 8 near-haploid and 4 low-hypodiploid B-ALL samples suggested that the massive loss of chromosomes is the primary oncogenic event, with other oncogenic insults occurring after hypodiploidy [20]. This is consistent with similar analyses in high-hyperdiploid B-ALL cases, the most frequent aneuploid entity in B-ALL, indicating that chromosome gains were the primary oncogenic event [37][38]. Thus, similar pathogenic mechanisms involving gross aneuploidies may be shared in these B-ALL subtypes. Furthermore, the genomic landscape of near-haploid and low-hypodiploid B-ALL subtypes, as well as that of high-hyperdiploid subtypes, is characterized by aneuploidy and subtype-specific mutations, with significant fewer microdeletions and structural chromosomal rearrangements in comparison with other cytogenetic subtypes containing structural chromosomal reorganizations [19][37]. Collectively, these data strongly suggest that hypodiploidy has a direct impact on cell transformation and leukemogenesis rather than being solely a passenger event. The fact that severe hypodiploidy is observed in a wide spectrum of neoplasms further indicates that it is indeed a major contributor of tumorigenesis [39].
6. Outcome and Treatment Strategies for B-ALL with Hypodiploidies <40 Chromosomes
6.1. Relationship of Genetic and Clinical Features with Patient Outcome
The EFS is not significantly different between patients with near-haploid or low-hypodiploid B-ALL, including those cases with “masked hypodiploidy” [13][14][35]. In some cases, hypodiploidy may accompany other primary genetic abnormalities, such as BCR-ABL1, TCF3-PBX1, ETV6-RUNX1 and KMT2A rearrangements, which modulate the prognosis of the disease. Accordingly, some authors have suggested that these patients should be treated based on the primary structural abnormalities rather than the hypodiploidy, and on their MRD values after induction [14]. The high presence of germline TP53 mutations among patients with low-hypodiploidy confer an increased risk of relapse in this group and is associated with the development of secondary neoplasms [14]. Therefore, it is highly recommended that all patients with low-hypodiploidy B-ALL are tested for germline TP53 mutations [14][40]. Strikingly, the germline TP53 mutations in these cases have been associated with increased mortality due to second neoplastic malignancies following hematopoietic stem cell transplantation (HSCT), highlighting the importance of the germline study in low-hypodiploid B-ALL to assess HSCT versus less toxic alternative therapies [16][41][42].
6.2. Current Treatment Protocols
Different study groups, such as the UKALL, NOPHO, AALL0031 and COG studies, consistently stratify near-haploid and low-hypodiploid B-ALL subtypes as high-risk based on the poor prognosis of the patients, which does not depend on treatment era or on the NCI risk group in which they are classified [14][16][43]. In view of the poor prognosis of patients with hypodiploid B-ALL, they have been classically treated with high-dose chemotherapy followed by allogeneic transplantation. However, different studies assessing the impact of HSCT on B-ALL with near-haploidy and low-hypodiploidy failed to demonstrate a clear benefit of HSCT in MRD positive or negative patients [13][14][35][44]. Notwithstanding these findings, the outcome of hypodiploid B-ALL with <40 chromosomes has been substantially improved by MRD-guided therapy, which intensifies treatments based on the MRD EOI status [13].
6.3. Novel Therapeutic Targets and Approaches to Treat B-ALL with <40 Chromosomes
New treatments aiming to target recently identified biological drivers of hypodiploidies as well as immunotherapy strategies are currently being explored to achieve better responses before HSCT or to be used as alternative approaches. The recent discovery of near-universal TP53 alterations in low-hypodiploid B-ALL has highlighted a key role for this gene in leukemogenesis. Investigation of this germinal mutation in this population is recommended when evaluating treatment with chemotherapy and HSCT. It remains to be demonstrated, however, whether therapies directed at this genetic lesion have an effect on low-hypodiploidy [36]. The anti-apoptotic protein BCL-2, has been identified as an effective therapeutic target for hypodiploid B-ALL with <40 chromosomes [45], and the efficacy of BCL-2 inhibitors (mainly venetoclax) has been demonstrated in ex vivo models of B-ALL with near-haploidy and low-hypodiploidy, especially in cases with elevated levels of the apoptosis-related factors BIM or BAD.
References
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390.
- Howlader, N.; Noone, A.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.; et al. SEER Cancer Statistics Review (CSR), 1975–2018. Available online: https://seer.cancer.gov/csr/1975_2018/ (accessed on 5 November 2021).
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30.
- Maitra, A.; McKenna, R.W.; Weinberg, A.G.; Schneider, N.R.; Kroft, S.H. Precursor B-cell lymphoblastic lymphoma. A study of nine cases lacking blood and bone marrow involvement and review of the literature. Am. J. Clin. Pathol. 2001, 115, 868–875.
- Molina, O.; Abad, M.A.; Sole, F.; Menendez, P. Aneuploidy in Cancer: Lessons from Acute Lymphoblastic Leukemia. Trends Cancer 2021, 7, 37–47.
- Pui, C.H.; Williams, D.L.; Raimondi, S.C.; Rivera, G.K.; Look, A.T.; Dodge, R.K.; George, S.L.; Behm, F.G.; Crist, W.M.; Murphy, S.B. Hypodiploidy is associated with a poor prognosis in childhood acute lymphoblastic leukemia. Blood 1987, 70, 247–253.
- Pui, C.H.; Carroll, A.J.; Raimondi, S.C.; Land, V.J.; Crist, W.M.; Shuster, J.J.; Williams, D.L.; Pullen, D.J.; Borowitz, M.J.; Behm, F.G.; et al. Clinical presentation, karyotypic characterization, and treatment outcome of childhood acute lymphoblastic leukemia with a near-haploid or hypodiploid less than 45 line. Blood 1990, 75, 1170–1177.
- Chessels, J.M.; Swansbury, G.J.; Reeves, B.; Bailey, C.C.; Richards, S.M. Cytogenetics and prognosis in childhood lymphoblastic leukaemia: Results of MRC UKALL X. Medical Research Council Working Party in Childhood Leukaemia. Br. J. Haematol. 1997, 99, 93–100.
- Heerema, N.A.; Nachman, J.B.; Sather, H.N.; Sensel, M.G.; Lee, M.K.; Hutchinson, R.; Lange, B.J.; Steinherz, P.G.; Bostrom, B.; Gaynon, P.S.; et al. Hypodiploidy with less than 45 chromosomes confers adverse risk in childhood acute lymphoblastic leukemia: A report from the children’s cancer group. Blood 1999, 94, 4036–4045.
- Raimondi, S.C.; Zhou, Y.; Mathew, S.; Shurtleff, S.A.; Sandlund, J.T.; Rivera, G.K.; Behm, F.G.; Pui, C.H. Reassessment of the prognostic significance of hypodiploidy in pediatric patients with acute lymphoblastic leukemia. Cancer 2003, 98, 2715–2722.
- Harrison, C.J.; Moorman, A.V.; Broadfield, Z.J.; Cheung, K.L.; Harris, R.L.; Reza Jalali, G.; Robinson, H.M.; Barber, K.E.; Richards, S.M.; Mitchell, C.D.; et al. Three distinct subgroups of hypodiploidy in acute lymphoblastic leukaemia. Br. J. Haematol. 2004, 125, 552–559.
- Nachman, J.B.; Heerema, N.A.; Sather, H.; Camitta, B.; Forestier, E.; Harrison, C.J.; Dastugue, N.; Schrappe, M.; Pui, C.H.; Basso, G.; et al. Outcome of treatment in children with hypodiploid acute lymphoblastic leukemia. Blood 2007, 110, 1112–1115.
- Mullighan, C.G.; Jeha, S.; Pei, D.; Payne-Turner, D.; Coustan-Smith, E.; Roberts, K.G.; Waanders, E.; Choi, J.K.; Ma, X.; Raimondi, S.C.; et al. Outcome of children with hypodiploid ALL treated with risk-directed therapy based on MRD levels. Blood 2015, 126, 2896–2899.
- Pui, C.H.; Rebora, P.; Schrappe, M.; Attarbaschi, A.; Baruchel, A.; Basso, G.; Cave, H.; Elitzur, S.; Koh, K.; Liu, H.C.; et al. Outcome of Children With Hypodiploid Acute Lymphoblastic Leukemia: A Retrospective Multinational Study. J. Clin. Oncol. 2019, 37, 770–779.
- Ishimaru, S.; Okamoto, Y.; Imai, C.; Sakaguchi, H.; Taki, T.; Hasegawa, D.; Cho, Y.; Kakuda, H.; Sano, H.; Manabe, A.; et al. Nationwide survey of pediatric hypodiploid acute lymphoblastic leukemia in Japan. Pediatr. Int. 2019, 61, 1103–1108.
- McNeer, J.L.; Devidas, M.; Dai, Y.; Carroll, A.J.; Heerema, N.A.; Gastier-Foster, J.M.; Kahwash, S.B.; Borowitz, M.J.; Wood, B.L.; Larsen, E.; et al. Hematopoietic Stem-Cell Transplantation Does Not Improve the Poor Outcome of Children With Hypodiploid Acute Lymphoblastic Leukemia: A Report From Children’s Oncology Group. J. Clin. Oncol. 2019, 37, 780–789.
- Safavi, S.; Paulsson, K. Near-haploid and low-hypodiploid acute lymphoblastic leukemia: Two distinct subtypes with consistently poor prognosis. Blood 2017, 129, 420–423.
- Mitelman, F.; Johansson, B.; Mertens, F. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. 2021. Available online: https://www.clinicalgenetics.lu.se/division-clinical-genetics/database-chromosome-aberrations-and-gene-fusions-cancer (accessed on 5 November 2021).
- Holmfeldt, L.; Wei, L.; Diaz-Flores, E.; Walsh, M.; Zhang, J.; Ding, L.; Payne-Turner, D.; Churchman, M.; Andersson, A.; Chen, S.C.; et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat. Genet. 2013, 45, 242–252.
- Safavi, S.; Forestier, E.; Golovleva, I.; Barbany, G.; Nord, K.H.; Moorman, A.V.; Harrison, C.J.; Johansson, B.; Paulsson, K. Loss of chromosomes is the primary event in near-haploid and low-hypodiploid acute lymphoblastic leukemia. Leukemia 2013, 27, 248–250.
- Muhlbacher, V.; Zenger, M.; Schnittger, S.; Weissmann, S.; Kunze, F.; Kohlmann, A.; Bellos, F.; Kern, W.; Haferlach, T.; Haferlach, C. Acute lymphoblastic leukemia with low hypodiploid/near triploid karyotype is a specific clinical entity and exhibits a very high TP53 mutation frequency of 93%. Genes Chromosomes Cancer 2014, 53, 524–536.
- Creasey, T.; Enshaei, A.; Nebral, K.; Schwab, C.; Watts, K.; Cuthbert, G.; Vora, A.; Moppett, J.; Harrison, C.J.; Fielding, A.K.; et al. Single nucleotide polymorphism array-based signature of low hypodiploidy in acute lymphoblastic leukemia. Genes Chromosomes Cancer 2021, 60, 604–615.
- Groupe Francais de Cytogenetique Hematologique. Cytogenetic abnormalities in adult acute lymphoblastic leukemia: Correlations with hematologic findings outcome. A Collaborative Study of the Group Francais de Cytogenetique Hematologique. Blood 1996, 87, 3135–3142.
- Charrin, C.; Thomas, X.; Ffrench, M.; Le, Q.H.; Andrieux, J.; Mozziconacci, M.J.; Lai, J.L.; Bilhou-Nabera, C.; Michaux, L.; Bernheim, A.; et al. A report from the LALA-94 and LALA-SA groups on hypodiploidy with 30 to 39 chromosomes and near-triploidy: 2 possible expressions of a sole entity conferring poor prognosis in adult acute lymphoblastic leukemia (ALL). Blood 2004, 104, 2444–2451.
- Pui, C.H.; Relling, M.V.; Downing, J.R. Acute lymphoblastic leukemia. N. Engl. J. Med. 2004, 350, 1535–1548.
- Carroll, A.J.; Shago, M.; Mikhail, F.M.; Raimondi, S.C.; Hirsch, B.A.; Loh, M.L.; Raetz, E.A.; Borowitz, M.J.; Wood, B.L.; Maloney, K.W.; et al. Masked hypodiploidy: Hypodiploid acute lymphoblastic leukemia (ALL) mimicking hyperdiploid ALL in children: A report from the Children’s Oncology Group. Cancer Genet. 2019, 238, 62–68.
- Harrison, C.J.; Moorman, A.V.; Barber, K.E.; Broadfield, Z.J.; Cheung, K.L.; Harris, R.L.; Jalali, G.R.; Robinson, H.M.; Strefford, J.C.; Stewart, A.; et al. Interphase molecular cytogenetic screening for chromosomal abnormalities of prognostic significance in childhood acute lymphoblastic leukaemia: A UK Cancer Cytogenetics Group Study. Br. J. Haematol. 2005, 129, 520–530.
- Ribera, J.; Granada, I.; Morgades, M.; Vives, S.; Genesca, E.; Gonzalez, C.; Nomdedeu, J.; Escoda, L.; Montesinos, P.; Mercadal, S.; et al. The poor prognosis of low hypodiploidy in adults with B-cell precursor acute lymphoblastic leukaemia is restricted to older adults and elderly patients. Br. J. Haematol. 2019, 186, 263–268.
- Stark, B.; Jeison, M.; Gobuzov, R.; Krug, H.; Glaser-Gabay, L.; Luria, D.; El-Hasid, R.; Harush, M.B.; Avrahami, G.; Fisher, S.; et al. Near haploid childhood acute lymphoblastic leukemia masked by hyperdiploid line: Detection by fluorescence in situ hybridization. Cancer Genet. Cytogenet. 2001, 128, 108–113.
- Safavi, S.; Olsson, L.; Biloglav, A.; Veerla, S.; Blendberg, M.; Tayebwa, J.; Behrendtz, M.; Castor, A.; Hansson, M.; Johansson, B.; et al. Genetic and epigenetic characterization of hypodiploid acute lymphoblastic leukemia. Oncotarget 2015, 6, 42793–42802.
- Agraz-Doblas, A.; Bueno, C.; Bashford-Rogers, R.; Roy, A.; Schneider, P.; Bardini, M.; Ballerini, P.; Cazzaniga, G.; Moreno, T.; Revilla, C.; et al. Unravelling the cellular origin and clinical prognostic markers of infant B-cell acute lymphoblastic leukemia using genome-wide analysis. Haematologica 2019, 104, 1176–1188.
- Ueno, H.; Yoshida, K.; Shiozawa, Y.; Nannya, Y.; Iijima-Yamashita, Y.; Kiyokawa, N.; Shiraishi, Y.; Chiba, K.; Tanaka, H.; Isobe, T.; et al. Landscape of driver mutations and their clinical impacts in pediatric B-cell precursor acute lymphoblastic leukemia. Blood Adv. 2020, 4, 5165–5173.
- Stengel, A.; Schnittger, S.; Weissmann, S.; Kuznia, S.; Kern, W.; Kohlmann, A.; Haferlach, T.; Haferlach, C. TP53 mutations occur in 15.7% of ALL and are associated with MYC-rearrangement, low hypodiploidy, and a poor prognosis. Blood 2014, 124, 251–258.
- Moorman, A.V. Does TP53 guard ALL genomes? Blood 2014, 124, 160–161.
- Miller, L.; Kobayashi, S.; Pauly, M.; Lew, G.; Saxe, D.; Keller, F.; Qayed, M.; Castellino, S. Evaluating approaches to enhance survival in children with hypodiploid acute lymphoblastic leukaemia (ALL). Br. J. Haematol. 2019, 185, 613–616.
- Comeaux, E.Q.; Mullighan, C.G. TP53 Mutations in Hypodiploid Acute Lymphoblastic Leukemia. Cold Spring Harb. Perspect. Med. 2017, 7, a026286.
- Paulsson, K.; Lilljebjorn, H.; Biloglav, A.; Olsson, L.; Rissler, M.; Castor, A.; Barbany, G.; Fogelstrand, L.; Nordgren, A.; Sjogren, H.; et al. The genomic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Nat. Genet. 2015, 47, 672–676.
- Paulsson, K. High hyperdiploid childhood acute lymphoblastic leukemia: Chromosomal gains as the main driver event. Mol. Cell. Oncol. 2016, 3, e1064555.
- Mandahl, N.; Johansson, B.; Mertens, F.; Mitelman, F. Disease-associated patterns of disomic chromosomes in hyperhaploid neoplasms. Genes Chromosomes Cancer 2012, 51, 536–544.
- Shetty, D.; Amare, P.K.; Mohanty, P.; Talker, E.; Chaubal, K.; Jain, H.; Tembhare, P.; Patkar, N.; Chaturvedi, A.; Subramanian, P.G.; et al. Investigating the clinical, hematological and cytogenetic profile of endoreduplicated hypodiploids in BCP-ALL. Blood Cells Mol. Dis. 2020, 85, 102465.
- Mehta, P.A.; Zhang, M.J.; Eapen, M.; He, W.; Seber, A.; Gibson, B.; Camitta, B.M.; Kitko, C.L.; Dvorak, C.C.; Nemecek, E.R.; et al. Transplantation Outcomes for Children with Hypodiploid Acute Lymphoblastic Leukemia. Biol. Blood Marrow Transplant. 2015, 21, 1273–1277.
- Winter, G.; Kirschner-Schwabe, R.; Groeneveld-Krentz, S.; Escherich, G.; Moricke, A.; von Stackelberg, A.; Stanulla, M.; Bailey, S.; Richter, L.; Steinemann, D.; et al. Clinical and genetic characteristics of children with acute lymphoblastic leukemia and Li-Fraumeni syndrome. Leukemia 2021, 35, 1475–1479.
- Lee, S.H.R.; Li, Z.; Tai, S.T.; Oh, B.L.Z.; Yeoh, A.E.J. Genetic Alterations in Childhood Acute Lymphoblastic Leukemia: Interactions with Clinical Features and Treatment Response. Cancers 2021, 13, 4068.
- Schultz, K.R.; Devidas, M.; Bowman, W.P.; Aledo, A.; Slayton, W.B.; Sather, H.; Zheng, H.W.; Davies, S.M.; Gaynon, P.S.; Trigg, M.; et al. Philadelphia chromosome-negative very high-risk acute lymphoblastic leukemia in children and adolescents: Results from Children’s Oncology Group Study AALL0031. Leukemia 2014, 28, 964–967.
- Diaz-Flores, E.; Comeaux, E.Q.; Kim, K.L.; Melnik, E.; Beckman, K.; Davis, K.L.; Wu, K.; Akutagawa, J.; Bridges, O.; Marino, R.; et al. Bcl-2 Is a Therapeutic Target for Hypodiploid B-Lineage Acute Lymphoblastic Leukemia. Cancer Res. 2019, 79, 2339–2351.
More
Information
Subjects:
Hematology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.6K
Revisions:
2 times
(View History)
Update Date:
10 Jan 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




