
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jesus Rodriguez Diaz | + 2207 word(s) | 2207 | 2021-12-21 09:52:53 | | | |
| 2 | Lindsay Dong | Meta information modification | 2207 | 2021-12-23 03:31:08 | | |
Video Upload Options
Rotavirus (RV) and norovirus (NoV) are the leading causes of acute gastroenteritis (AGE) worldwide. Histo-blood group antigens (HBGAs) have a role in NoV and RV infections since their presence on the gut epithelial surfaces is essential for the susceptibility to many NoV and RV genotypes. A second factor that influences enteric viral infections is the gut microbiota of the host. In vitro and animal studies have determined that the gut microbiota limits, but in some cases enhances enteric viral infection. The ways that microbiota can enhance NoV or RV infection include virion stabilization and promotion of virus attachment to host cells, whereas experiments with microbiota-depleted and germ-free animals point to immunoregulation as the mechanism by which the microbiota restrict infection.
1. Enteric Viruses and Their Impact on Human Health
2. Glycobiology Mediates Enteric Virus/Host Interactions
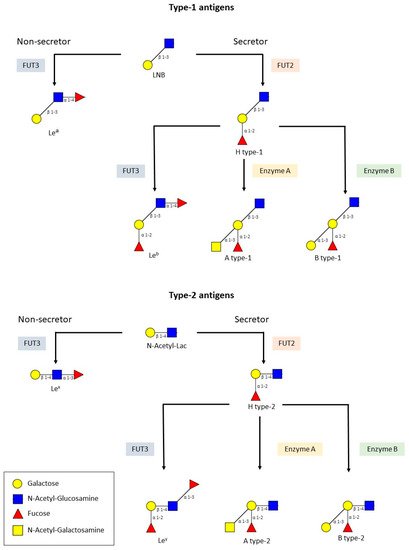
2.1. HBGAs and RV
2.2. HBGAs and NoV
3. The Role of Bacteria in RVs and NoVs Infection
3.1. Bacteria against Enteric Viral Infections
3.2. Microbiota and Promotion of Enteric Viral Infections
3.3. Microbiota and Restriction of Enteric Viral Infections
4. Microbiota and Enteric Viruses, Studies in Humans
Microbiota composition varies depending on the population [41] since it is affected by many factors including nutrition [42], sex, age, genetics, and health status [43], and these vary greatly between low-income and high-income countries. Such differences could be some of the reasons why RVVs have significantly lower efficacy in low-income countries [44][45].
A recent study evaluated whether microbiota modification by the use of broad- and narrow-spectrum antibiotics had an effect on immunization with Rotarix in adults [46]. Although the experimental groups did not differ in terms of total IgA produced, the narrow spectrum group showed a boost in IgA at day seven post-vaccination (basal levels of anti-RV IgA were high in the vaccination group) and the viral shedding was increased in both groups treated with antibiotics. Differences in the microbiota composition in faeces were evident between the groups and correlations between enrichment in Bacteroides populations at the boost at day seven were observed and several taxa (Prevotellaceae, Cloacibacillus everynsis, and Proteobacteria members such as Escherichia and Shigella) were associated with increased viral shedding. Antibiotic treatment had no effect on the immunogenicity of other systemic vaccines applied (pneumococcal and tetanus vaccine) [46]. These results highlight the fact that targeting the microbiota could be an alternative strategy to enhance RVVs efficacy, although the effectiveness in children still needs further investigation.
References
- World Health Organization (WHO). The Top 10 Causes of Death. Available online: http://www.who.int/en/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 10 September 2021).
- Donaldson, E.F.; Lindesmith, L.C.; Lobue, A.D.; Baric, R.S. Norovirus pathogenesis: Mechanisms of persistence and immune evasion in human populations. Immunol. Rev. 2008, 225, 190–211.
- Jiang, X.; Liu, Y.; Tan, M. Histo-blood group antigens as receptors for rotavirus, new understanding on rotavirus epidemiology and vaccine strategy. Emerg. Microbes Infect. 2017, 6, e22.
- Shanker, S.; Czakó, R.; Sapparapu, G.; Alvarado, G.; Viskovska, M.; Sankaran, B.; Atmar, R.L.; Crowe, J.E.; Estes, M.K.; Prasad, B.V.V. Structural basis for norovirus neutralization by an HBGA blocking human IgA antibody. Proc. Natl. Acad. Sci. USA 2016, 113, E5830–E5837.
- Marionneau, S.; Cailleau-Thomas, A.; Rocher, J.; Le Moullac-Vaidye, B.; Ruvoën, N.; Clément, M.; Le Pendu, J. ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie 2001, 83, 565–573.
- Tan, M.; Jiang, X. Histo-blood group antigens: A common niche for norovirus and rotavirus. Expert. Rev. Mol. Med. 2014, 16, e5.
- Barbé, L.; Le Pendu, J.; Echasserieau, K.; Ruvoën-Clouet, N.; Bernardeau, K.; Le Moullac-Vaidye, B.; Bovin, N.; Nordgren, J.; Carton, T.; Svensson, L. Histo-blood group antigen-binding specificities of human rotaviruses are associated with gastroenteritis but not with in vitro infection. Sci. Rep. 2018, 8, 12961.
- Marionneau, S.; Ruvoën, N.; Le MoullacVaidye, B.; Clement, M.; CailleauThomas, A.; RuizPalacois, G.; Huang, P.; Jiang, X.; Le Pendu, J. Norwalk Virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 2002, 122, 1967–1977.
- Ayouni, S.; Sdiri-Loulizi, K.; de Rougemont, A.; Estienney, M.; Ambert-Balay, K.; Aho, S.; Hamami, S.; Aouni, M.; Neji-Guediche, M.; Pothier, P.; et al. Rotavirus P infections in persons with secretor and nonsecretor phenotypes, Tunisia. Emerg. Infect. Dis. 2015, 21, 2055–2058.
- Arias, C.F.; López, S. Rotavirus cell entry: Not so simple after all. Curr. Opin. Virol. 2021, 48, 42–48.
- Böhm, R.; Fleming, F.E.; Maggioni, A.; Dang, V.T.; Holloway, G.; Coulson, B.S.; Von Itzstein, M.; Haselhorst, T. Revisiting the role of histo-blood group antigens in rotavirus host-cell invasion. Nat. Commun. 2015, 6, 5907.
- Liu, Y.Y.; Huang, P.; Tan, M.; Biesiada, J.; Meller, J.; Castello, A.A.; Jiang, B.; Jiang, X. Rotavirus VP8*: Phylogeny, host range, and interaction with histo-blood group antigens. J. Virol. 2012, 86, 9899–9910.
- Xu, S.; McGinnis, K.R.; Liu, Y.; Huang, P.; Tan, M.; Stuckert, M.R.; Burnside, R.E.; Jacob, E.G.; Ni, S.; Jiang, X.; et al. Structural basis of P rotavirus evolution and host ranges under selection of histo-blood group antigens. Proc. Natl. Acad. Sci. USA 2021, 118, e2107963118.
- Huang, P.; Xia, M.; Tan, M.; Zhong, W.; Wei, C.; Wang, L.; Morrow, A.; Jiang, X. Spike protein VP8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. J. Virol. 2012, 86, 4833–4843.
- Xu, S.; Ahmed, L.U.; Stuckert, M.R.; McGinnis, K.R.; Liu, Y.; Tan, M.; Huang, P.; Zhong, W.; Zhao, D.; Jiang, X.; et al. Molecular basis of P major human rotavirus VP8* domain recognition of histo-blood group antigens. PLoS Pathog. 2020, 16, e1008386.
- Fix, J.; Chandrashekhar, K.; Perez, J.; Bucardo, F.; Hudgens, M.G.; Yuan, L.; Twitchell, E.; Azcarate-Peril, M.A.; Vilchez, S.; Becker-Dreps, S. Association between Gut Microbiome Composition and Rotavirus Vaccine Response among Nicaraguan Infants. Am. J. Trop. Med. Hyg. 2020, 102, 213–219.
- Gozalbo-Rovira, R.; Ciges-Tomas, J.R.; Vila-Vicent, S.; Buesa, J.; Santiso-Bellón, C.; Monedero, V.; Yebra, M.J.; Marina, A.; Rodríguez-Díaz, J. Unraveling the role of the secretor antigen in human rotavirus attachment to histo-blood group antigens. PLoS Pathog. 2019, 15, e1007865.
- Liu, Y.; Xu, S.; Woodruff, A.L.; Xia, M.; Tan, M.; Kennedy, M.A.; Jiang, X. Structural basis of glycan specificity of P VP8*: Implications for rotavirus zoonosis and evolution. PLoS Pathog. 2017, 13, e1006707.
- Lee, S.-K.; Oh, S.J.; Choi, S.; Choi, S.H.; Shin, S.-H.; Lee, E.J.; Cho, E.-J.; Hyun, J.; Kim, H.S. Relationship Between Rotavirus P Infection in Korean Neonates and Histo-Blood Group Antigen: A Single-Center Study. Ann. Lab. Med. 2021, 41, 181–189.
- Liu, Y.; Ramelot, T.A.; Huang, P.; Liu, Y.; Li, Z.; Feizi, T.; Zhong, W.; Wu, F.-T.; Tan, M.; Kennedy, M.A.; et al. Glycan Specificity of P Rotavirus and Comparison with Those of Related P Genotypes. J. Virol. 2016, 90, 9983–9996.
- Hu, L.; Crawford, S.E.; Czako, R.; Cortes-Penfield, N.W.; Smith, D.F.; Le Pendu, J.; Estes, M.K.; Prasad, B.V.V. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature 2012, 485, 256–259.
- Liu, Y.; Huang, P.; Jiang, B.; Tan, M.; Morrow, A.L.; Jiang, X. Poly-LacNAc as an Age-Specific Ligand for Rotavirus P in Neonates and Infants. PLoS ONE 2013, 8, e78113.
- Hu, L.; Sankaran, B.; Laucirica, D.R.; Patil, K.; Salmen, W.; Ferreon, A.C.M.; Tsoi, P.S.; Lasanajak, Y.; Smith, D.F.; Ramani, S.; et al. Glycan recognition in globally dominant human rotaviruses. Nat. Commun. 2018, 9, 2631.
- Huang, P.; Farkas, T.; Zhong, W.; Tan, M.; Thornton, S.; Morrow, A.L.; Jiang, X. Norovirus and Histo-Blood Group Antigens: Demonstration of a Wide Spectrum of Strain Specificities and Classification of Two Major Binding Groups among Multiple Binding Patterns. J. Virol. 2005, 79, 6714–6722.
- Carmona-Vicente, N.; Vila-Vicent, S.; Allen, D.; Gozalbo-Rovira, R.; Iturriza-Gómara, M.; Buesa, J.; Rodríguez-Díaz, J. Characterization of a Novel Conformational GII.4 Norovirus Epitope: Implications for Norovirus-Host Interactions. J. Virol. 2016, 90, 7703–7714.
- Almand, E.A.; Moore, M.D.; Jaykus, L.-A. Norovirus Binding to Ligands Beyond Histo-Blood Group Antigens. Front. Microbiol. 2017, 8, 2549.
- Zheng, L.; Zhang, H.; Ma, J.; Liu, J.; Ma, S.; Wang, M.; Huo, Y. Phylogenetic and biological characterizations of a GI.3 norovirus. Infect. Genet. Evol. 2020, 85, 104554.
- Hao, Q.; Lu, Z.; Dong, B.R.; Huang, C.Q.; Wu, T. Probiotics for preventing acute upper respiratory tract infections. Cochrane Database Syst. Rev. 2015, 2, CD006895.
- Ahmadi, E.; Alizadeh-Navaei, R.; Rezai, M.S. Efficacy of probiotic use in acute rotavirus diarrhea in children: A systematic review and meta-analysis. Casp. J. Intern. Med. 2015, 6, 187–195.
- Wu, Y.; Zhang, Q.; Ren, Y.; Ruan, Z. Effect of probiotic Lactobacillus on lipid profile: A systematic review and meta-analysis of randomized, controlled trials. PLoS ONE 2017, 12, e0178868.
- Rees, C.M.; Hall, N.J.; Fleming, P.; Eaton, S. Probiotics for the prevention of surgical necrotising enterocolitis: Systematic review and meta-analysis. BMJ Paediatr. Open 2017, 1, e000066.
- Lei, S.; Twitchell, E.; Yuan, L. Pathogenesis, Immunity and the Role of Microbiome/Probiotics in Enteric Virus Infections in Humans and Animal Models. In Mechanisms Underlying Host-Microbiome Interactions in Pathophysiology of Human Diseases; Springer: Boston, MA, USA, 2018; pp. 55–78.
- Monedero, V.; Rodríguez-Díaz, J. Intestinal microbiota and susceptibility to viral infections: Role of probiotics. Probiotics Prebiotics Synbiotics 2016, 813–826.
- Domínguez-Díaz, C.; García-Orozco, A.; Riera-Leal, A.; Padilla-Arellano, J.R.; Fafutis-Morris, M. Microbiota and Its Role on Viral Evasion: Is It with Us or Against Us? Front. Cell. Infect. Microbiol. 2019, 9, 256.
- Kuss, S.K.; Best, G.T.; Etheredge, C.A.; Pruijssers, A.J.; Frierson, J.M.; Hooper, L.V.; Dermody, T.S.; Pfeiffer, J.K. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 2011, 334, 249–252.
- Kane, M.; Case, L.K.; Kopaskie, K.; Kozlova, A.; MacDearmid, C.; Chervonsky, A.V.; Golovkina, T.V. Successful Transmission of a Retrovirus Depends on the Commensal Microbiota. Science 2011, 334, 245–249.
- Uchiyama, R.; Chassaing, B.; Zhang, B.; Gewirtz, A.T. Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J. Infect. Dis. 2014, 210, 171–182.
- Jones, M.K.; Watanabe, M.; Zhu, S.; Graves, C.L.; Keyes, L.R.; Grau, K.R.; Gonzalez-Hernandez, M.B.; Iovine, N.M.; Wobus, C.E.; Vinje, J.; et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 2014, 346, 755–759.
- Gozalbo-Rovira, R.; Santiso-Bellón, C.; Buesa, J.; del Campo, A.R.; Vila-Vicent, S.; Muñoz, C.; Yebra, M.J.; Monedero, V.; Rodríguez-Díaz, J. Microbiota Depletion Promotes Human Rotavirus Replication in an Adult Mouse Model. Biomedicines 2021, 9, 846.
- Schnepf, D.; Hernandez, P.; Mahlakõiv, T.; Crotta, S.; Sullender, M.E.; Peterson, S.T.; Ohnemus, A.; Michiels, C.; Gentle, I.; Dumoutier, L.; et al. Rotavirus susceptibility of antibiotic-treated mice ascribed to diminished expression of interleukin-22. PLoS ONE 2021, 16, e0247738.
- Gupta, V.K.; Paul, S.; Dutta, C. Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity. Front. Microbiol. 2017, 8, 1162.
- Srivastava, V.; Deblais, L.; Huang, H.-C.; Miyazaki, A.; Kandasamy, S.; Langel, S.N.; Paim, F.C.; Chepngeno, J.; Kathayat, D.; Vlasova, A.N.; et al. Reduced rotavirus vaccine efficacy in protein malnourished human-faecal-microbiota-transplanted gnotobiotic pig model is in part attributed to the gut microbiota. Benef. Microbes 2020, 11, 733–751.
- Desselberger, U. Differences of Rotavirus Vaccine Effectiveness by Country: Likely Causes and Contributing Factors. Pathogens 2017, 6, 65.
- Armah, G.E.; Sow, S.O.; Breiman, R.F.; Dallas, M.J.; Tapia, M.D.; Feikin, D.R.; Binka, F.N.; Steele, A.D.; Laserson, K.F.; Ansah, N.A.; et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-Saharan Africa: A randomised, double-blind, placebo-controlled trial. Lancet 2010, 376, 606–614.
- Madhi, S.A.; Cunliffe, N.A.; Steele, D.; Witte, D.; Kirsten, M.; Louw, C.; Ngwira, B.; Victor, J.C.; Gillard, P.H.; Cheuvart, B.B.; et al. Effect of Human Rotavirus Vaccine on Severe Diarrhea in African Infants. N. Engl. J. Med. 2010, 362, 289–298.
- Harris, V.C.; Haak, B.W.; Handley, S.A.; Jiang, B.; Velasquez, D.E.; Hykes, B.L.; Droit, L.; Berbers, G.A.M.; Kemper, E.M.; van Leeuwen, E.M.M.; et al. Effect of Antibiotic-Mediated Microbiome Modulation on Rotavirus Vaccine Immunogenicity: A Human, Randomized-Control Proof-of-Concept Trial. Cell Host Microbe 2018, 24, 197–207.e4.
- Gozalbo-Rovira, R.; Rubio-Del-campo, A.; Santiso-Bellón, C.; Vila-Vicent, S.; Buesa, J.; Delgado, S.; Molinero, N.; Margolles, A.; Yebra, M.J.; Collado, M.C.; et al. Interaction of intestinal bacteria with human rotavirus during infection in children. Int. J. Mol. Sci. 2021, 22, 1010.
- Dinleyici, E.C.; Martínez-Martínez, D.; Kara, A.; Karbuz, A.; Dalgic, N.; Metin, O.; Yazar, A.S.; Guven, S.; Kurugol, Z.; Turel, O.; et al. Time Series Analysis of the Microbiota of Children Suffering from Acute Infectious Diarrhea and Their Recovery After Treatment. Front. Microbiol. 2018, 9, 1230.




