
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Hilda Esperanza Esparza-Ponce | + 2221 word(s) | 2221 | 2021-12-06 10:49:30 | | | |
| 2 | Peter Tang | Meta information modification | 2221 | 2021-12-18 07:41:45 | | |
Video Upload Options
A bacterium becomes resistant due to the transfer of genes encoding antibiotic resistance. Bacteria constantly mutate; therefore, their defense mechanisms change constantly. Nanotechnology plays a key role in antimicrobial resistance due to materials modified at the nanometer scale, allowing large numbers of molecules to assemble to have a dynamic interface. These nanomaterials act as carriers, and their design is mainly focused on introducing the temporal and spatial release of the payload of antibiotics. In addition, they generate new antimicrobial modalities for the bacteria, which are not capable of protecting themselves.
1. Introduction
|
Priority |
Pathogenic Bacteria |
Antibiotics for Which There is Resistance |
|---|---|---|
|
Critical |
Acinetobacter baumannii |
Carbapenem |
|
Pseudomonas aeruginosa |
||
|
Enterobacteriaceae |
||
|
Mycobacteria |
Carbapenem and 3rd generation cephalosporins |
|
|
Mycobacterium tuberculosis |
3rd generation cephalosporins |
|
|
High |
Enterococcus faecium |
Vancomycin and methicillin |
|
Staphylococcus aureus |
||
|
Helicobacter pylori |
Vancomycin |
|
|
Campylobacter |
Clarithromycin |
|
|
Salmonella spp. |
Fluoroquinolones |
|
|
Neisseria gonorrhoeae |
3rd generation fluoroquinolone |
|
|
Medium |
Streptococcus pneumoniae |
|
|
Haemophilus influenza |
Non-sensible to penicillin |
|
|
Shigella spp. |
Ampicillin and fluroquinolones |
2. Antibiotics
2.1. By Chemical Structure
2.2. By Effect
2.3. By Spectrum
2.4. By the Mechanism of Action
3. Antimicrobial Resistance
4. Nanotechnology Applied to Antimicrobial Resistance
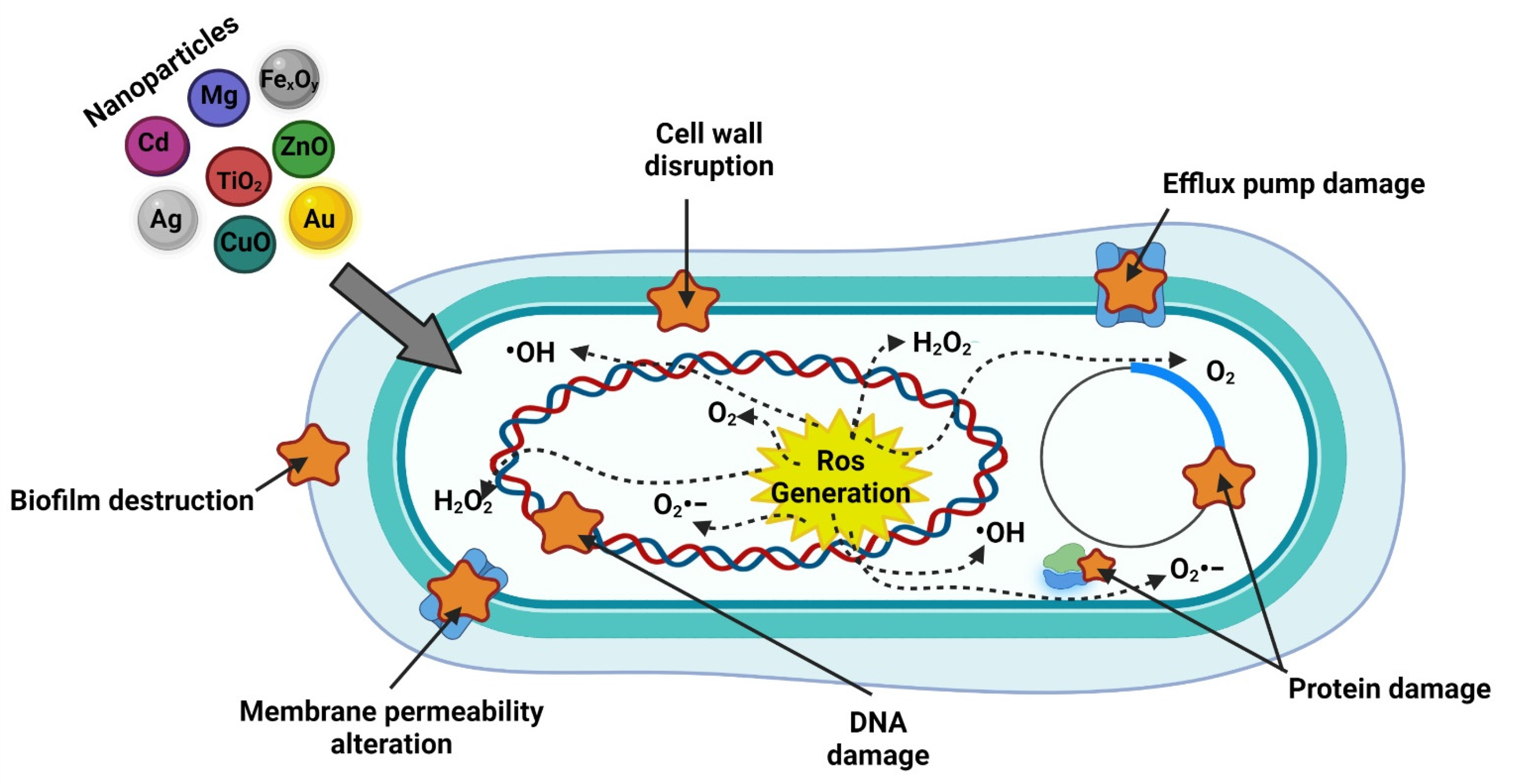
References
- Econom, D.E. NORMA MEXICANA NMX-R-80004-1-SCFI-2013 Nanotecnologías—Vocabulario—Parte 1: Conceptos Básicos Nanotechnologies—Vocabulary—Part 1: Core Terms, Dirección General de Normas (DGN); Dirección General de Normas (DGN): Cuauhtémoc, México, 2013.
- Bayda, S.; Adeel, M.; Tuccinardi, T.; Cordani, M.; Rizzolio, F.; Baeza, A. The History of Nanoscience and Nanotechnology: From Chemical-Physical Applications to Nanomedicine. Molecules 2020, 25, 112.
- Kandi, V.; Kandi, S. Antimicrobial properties of nanomolecules: Potential candidates as antibiotics in the era of multi-drug resistance. Epidemiol. Health 2015, 37, e2015020.
- Salomoni, R.; Léo, P.; Montemor, A.; Rinaldi, B.; Rodrigues, M. Antibacterial effect of silver nanoparticles in Pseudomonas aeruginosa. Nanotechnol. Sci. Appl. 2017, 10, 115–121.
- NNI. Nanotechnology. Available online: www.nano.gov (accessed on 15 March 2021).
- Yougbare, S.; Chang, T.-K.; Tan, S.-H.; Kuo, J.-C.; Hsu, P.-H.; Su, C.-Y.; Kuo, T.-R. Antimicrobial Gold Nanoclusters: Recent Developments and Future Perspectives. Int. J. Mol. Sci. 2019, 20, 2924.
- Contera, S.; Bernardino de la Serna, J.; Tetley, T.D. Biotechnology, nanotechnology and medicine. Emerg. Top. Life Sci. 2020, 4, 551–554.
- Allahverdiyev, A.M.; Kon, K.V.; Abamor, E.S.; Bagirova, M.; Rafailovich, M. Coping with antibiotic resistance: Combining nanoparticles with antibiotics and other antimicrobial agents. Expert Rev. Anti. Infect. Ther. 2011, 9, 1035–1052.
- Roy, J.; Chandra, S.; Maitra, S. Nanotechnology in castable refractory. Ceram. Int. 2019, 45, 19–29.
- Peigneux, A.; Oltolina, F.; Colangelo, D.; Iglesias, G.R.; Delgado, A.V.; Prat, M.; Jimenez-Lopez, C. Functionalized Biomimetic Magnetic Nanoparticles as Effective Nanocarriers for Targeted Chemotherapy. Part. Part. Syst. Charact. 2019, 36, 1900057.
- Kuo, Y.-L.; Wang, S.-G.; Wu, C.-Y.; Lee, K.-C.; Jao, C.-J.; Chou, S.-H.; Chen, Y.-C. Functional gold nanoparticle-based antibacterial agents for nosocomial and antibiotic-resistant bacteria. Nanomedicine 2016, 11, 2497–2510.
- Kumari, A.; Singla, R.; Guliani, A.; Yadav, S.K. Nanoencapsulation for drug delivery. EXCLI J. 2014, 13, 265–286.
- Menon, S.; Rajeshkumar, S.; Kumar, V. A review on biogenic synthesis of gold nanoparticles, characterization, and its applications. Resour. Technol. 2017, 3, 516–527.
- Kim, B.Y.S.; Rutka, J.T.; Chan, W.C.W. Nanomedicine. N. Engl. J. Med. 2010, 363, 2434–2443.
- Blecher, K.; Nasir, A.; Friedman, A. The growing role of nanotechnology in combating infectious disease. Virulence 2011, 2, 395–401.
- Ratner, B.D. Biomaterials: Been There, Done That, and Evolving into the Future. Annu. Rev. Biomed. Eng. 2019, 21, 171–191.
- OMS. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Available online: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf?ua=1 (accessed on 25 February 2020).
- Li, B.; Webster, T.J. Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopedic infections. J. Orthop. Res. 2017, 36, 22–32.
- Kumar, L.; Verma, S.; Vaidya, B.; Mehra, N.K. Nanocarrier-Assisted Antimicrobial Therapy against Intracellular Pathogens; Chapter 13; Elsevier: Amsterdam, The Netherlands, 2017; pp. 293–324. ISBN 9780323461528.
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80.
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.M.; Wertheim, H.F.L.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098.
- Feng, Q.; Huang, Y.; Chen, M.; Li, G.; Chen, Y. Functional synergy of α-helical antimicrobial peptides and traditional antibiotics against Gram-negative and Gram-positive bacteria in vitro and in vivo. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 197–204.
- CDC. Antimicrobial Resistance. Available online: https://www.cdc.gov/drugresistance/biggest-threats.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fdrugresistance%2Fbiggest_threats.html (accessed on 15 March 2021).
- CDC. Antibiotic Resistance Threats in the United States; U.S. Centers for Disease Control and Prevention: Atlanta, GA, USA, 2019.
- Eyler, R.F.; Shvets, K. Clinical Pharmacology of Antibiotics. Clin. J. Am. Soc. Nephrol. 2019, 14, 1080–1090.
- Brunton, L.; Hilal-Dandan, R.; Knollmann BCMacDougall, C. Penicillins, cephalosporins and other β-lactam antibiotics. In Goodman & Gilman’s: The Pharmacological Basis of Therapeutics; Brunton, L., Hilal-Dandan, R., BC, K., Eds.; McGraw-Hill Education: New York, NY, USA, 2017; pp. 1023–1038.
- Lodise, T.P.; Lomaestro, B.; Drusano, G.L. Piperacillin-Tazobactam for Pseudomonas aeruginosa Infection: Clinical Implications of an Extended-Infusion Dosing Strategy. Clin. Infect. Dis. 2007, 44, 357–363.
- Koomanachai, P.; Bulik, C.C.; Kuti, J.L.; Nicolau, D.P. Pharmacodynamic modeling of intravenous antibiotics against gram-negative bacteria collected in the United States. Clin. Ther. 2010, 32, 766–779.
- Lingzhi, L.; Haojie, G.; Dan, G.; Hongmei, M.; Yang, L.; Mengdie, J.; Chengkun, Z.; Xiaohui, Z. The role of two-component regulatory system in β-lactam antibiotics resistance. Microbiol. Res. 2018, 215, 126–129.
- Werth, B.J. Overview of Antibiotics. Available online: https://www.msdmanuals.com/home/infections/antibiotics/overview-of-antibiotics (accessed on 3 March 2020).
- Murray, P.R.; Rosenthal, K.S.; Pfaller, M.A. Bacteriology. In Medical Microbiology; Saunders, E., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; p. 872. ISBN 9780323673228.
- Brown, E.D.; Wright, G.D. Antibacterial drug discovery in the resistance era. Nature 2016, 529, 336–343.
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461.
- Banin, E.; Hughes, D.; Kuipers, O.P. Editorial: Bacterial pathogens, antibiotics and antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 450–452.
- Luepke, K.H.; Suda, K.J.; Boucher, H.; Russo, R.L.; Bonney, M.W.; Hunt, T.D.; Mohr, J.F. Past, Present, and Future of Antibacterial Economics: Increasing Bacterial Resistance, Limited Antibiotic Pipeline, and Societal Implications. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2017, 37, 71–84.
- Chung, P.Y.; Khanum, R. Antimicrobial peptides as potential anti-biofilm agents against multidrug-resistant bacteria. J. Microbiol. Immunol. Infect. 2017, 50, 405–410.
- Cárdenas, L.L.; Merchán, M.A.; López, D.P. New antibiotics against bacterial resistance. Infectio 2019, 23, 382.
- MacFadden, D.R.; McGough, S.F.; Fisman, D.; Santillana, M.; Brownstein, J.S. Antibiotic resistance increases with local temperature. Nat. Clim. Chang. 2018, 8, 510–514.
- Imran, M.; Das, K.R.; Naik, M.M. Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: An emerging health threat. Chemosphere 2019, 215, 846–857.
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658.
- Frieri, M.; Kumar, K.; Boutin, A. Antibiotic resistance. J. Infect. Public Health 2017, 10, 369–378.
- Troncoso, C.; Pavez, M.; Santos, A.; Salazar, R.; Barrientos, L. Implicancias Estructurales y Fisiológicas de la Célula Bacteriana en los Mecanismos de Resistencia Antibiótica. Int. J. Morphol. 2017, 35, 1214–1223.
- Bullington, W.; Hempstead, S.; Smyth, A.R.; Drevinek, P.; Saiman, L.; Waters, V.J.; Bell, S.C.; VanDevanter, D.R.; Flume, P.A.; Elborn, S.; et al. Antimicrobial resistance: Concerns of healthcare providers and people with CF. J. Cyst. Fibros. 2020, 20, 407–412.
- Płusa, T.; Konieczny, R.; Baranowska, A.; Szymczak, Z. The growing resistance of bacterial strains to antibiotics. Pol. Merkur. Lek. 2019, 47, 106–110.
- Mobarki, N.; Almerabi, B.; Hattan, A. Antibiotic Resistance Crisis. Int. J. Med. Dev. Ctries 2019, 561–564.
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482–501.
- Mandsberg, L.F.; Ciofu, O.; Kirkby, N.; Christiansen, L.E.; Poulsen, H.E.; Høiby, N. Antibiotic Resistance in Pseudomonas aeruginosa Strains with Increased Mutation Frequency Due to Inactivation of the DNA Oxidative Repair System. Antimicrob. Agents Chemother. 2009, 53, 2483–2491.
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and resistance mechanisms of antibiotics: A guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300.
- Ruddaraju, L.K.; Pammi, S.V.N.; Guntuku, G.S.; Padavala, V.S.; Kolapalli, V.R.M. A review on anti-bacterials to combat resistance: From ancient era of plants and metals to present and future perspectives of green nano technological combinations. Asian J. Pharm. Sci. 2020, 15, 42–59.
- Nas, F. Mechanisms of Bacterial Antibiotics Resistance: A Review. J. Adv. Microbiol. 2017, 7, 1–6.
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17.
- Majeed, H.T.; Aljanaby, A.A.J. Antibiotic Susceptibility Patterns and Prevalence of Some Extended Spectrum Beta-Lactamases Genes in Gram-Negative Bacteria Isolated from Patients Infected with Urinary Tract Infections in Al-Najaf City, Iraq. Avicenna J. Med. Biotechnol. 2019, 11, 192–201.
- Bassetti, M.; Vena, A.; Croxatto, A.; Righi, E.; Guery, B. How to manage Pseudomonas aeruginosa infections. Drugs Context 2018, 7, 1–18.
- Henrichfreise, B.; Wiegand, I.; Pfister, W.; Wiedemann, B. Resistance Mechanisms of Multiresistant Pseudomonas aeruginosa Strains from Germany and Correlation with Hypermutation. Antimicrob. Agents Chemother. 2007, 51, 4062–4070.
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51.
- Arzanlou, M.; Chai, W.C.; Venter, H. Intrinsic, adaptive and acquired antimicrobial resistance in Gram-negative bacteria. Essays Biochem. 2017, 61, 49–59.
- Yang, Y.; Mathieu, J.M.; Chattopadhyay, S.; Miller, J.T.; Wu, T.; Shibata, T.; Guo, W.; Alvarez, P.J.J. Defense Mechanisms of Pseudomonas aeruginosa PAO1 against Quantum Dots and Their Released Heavy Metals. ACS Nano 2012, 6, 6091–6098.
- Hancock, R.E.W.; Brinkman, F.S.L. Function of Pseudomonas Porins in Uptake and Efflux. Annu. Rev. Microbiol. 2002, 56, 17–38.
- Bradford, P.A. Epidemiology of Bacterial Resistance. In Antimicrobial Resistance in the 21st Century; Springer International Publishing: Cham, Switzerland, 2018; pp. 299–339. ISBN 978-3-319-78538-7.
- Bockstael, K.; Aerschot, A. Antimicrobial resistance in bacteria. Open Med. 2009, 4, 141–155.
- Hasan Abusaiba, T.; AL-Harmoosh, R. Mechanisms of Antibiotics Resistance in Bacteria. Syst. Rev. Pharm. 2020, 11, 817–823.
- Begum, S.; Begum, T.; Rahman, N.; Khan, R.A. A review on antibiotic resistance and way of combating antimicrobial resistance. GSC Biol. Pharm. Sci. 2021, 14, 087–097.
- Miller, K.P.; Wang, L.; Benicewicz, B.C.; Decho, A.W. Inorganic nanoparticles engineered to attack bacteria. Chem. Soc. Rev. 2015, 44, 7787–7807.
- Piñón-Castillo, H.A.; Martínez-Chamarro, R.; Reyes-Martínez, R.; Salinas-Vera, Y.M.; Manjarrez-Nevárez, L.A.; Muñoz-Castellanos, L.N.; López-Camarillo, C.; Orrantia-Borunda, E. Palladium Nanoparticles Functionalized with PVP-Quercetin Inhibits Cell Proliferation and Activates Apoptosis in Colorectal Cancer Cells. Appl. Sci. 2021, 11, 1988.
- Shrestha, A.; Kishen, A. Antibacterial Nanoparticles in Endodontics: A Review. J. Endod. 2016, 42, 1417–1426.
- Bi, J.; Li, T.; Ren, H.; Ling, R.; Wu, Z.; Qin, W. Capillary electrophoretic determination of heavy-metal ions using 11-mercaptoundecanoic acid and 6-mercapto-1-hexanol co-functionalized gold nanoparticle as colorimetric probe. J. Chromatogr. A 2019, 1594, 208–215.
- Thakkar, K.N.; Mhatre, S.S.; Parikh, R.Y. Biological synthesis of metallic nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2010, 6, 257–262.
- Brewer, M.; Zhang, T.; Dong, W.; Rutherford, M.; Tian, Z.R. Future Approaches of Nanomedicine in Clinical Science. Med. Clin. N. Am. 2007, 91, 963–1016.
- McNamara, K.; Tofail, S.A.M. Nanosystems: The use of nanoalloys, metallic, bimetallic, and magnetic nanoparticles in biomedical applications. Phys. Chem. Chem. Phys. 2015, 17, 27981–27995.
- Kaweeteerawat, C.; Na Ubol, P.; Sangmuang, S.; Aueviriyavit, S.; Maniratanachote, R. Mechanisms of antibiotic resistance in bacteria mediated by silver nanoparticles. J. Toxicol. Environ. Health Part. A 2017, 80, 1276–1289.
- Li, X.; Xu, H.; Chen, Z.-S.; Chen, G. Biosynthesis of Nanoparticles by Microorganisms and Their Applications. J. Nanomater. 2011, 2011, 270974.
- Liu, J.; Qiao, S.Z.; Hu, Q.H.; Max Lu, G.Q. Magnetic Nanocomposites with Mesoporous Structures: Synthesis and Applications. Small 2011, 7, 425–443.
- Spirescu, V.A.; Niculescu, A.-G.; Slave, Ș.; Bîrcă, A.C.; Dorcioman, G.; Grumezescu, V.; Holban, A.M.; Oprea, O.-C.; Vasile, B.Ș.; Grumezescu, A.M.; et al. Anti-Biofilm Coatings Based on Chitosan and Lysozyme Functionalized Magnetite Nanoparticles. Antibiotics 2021, 10, 1269.
- Motelica, L.; Ficai, D.; Oprea, O.; Ficai, A.; Trusca, R.D.; Andronescu, E.; Holban, A.M. Biodegradable alginate films with ZnO nanoparticles and citronella essential oil-a novel antimicrobial structure. Pharmaceutics 2021, 13, 1020.
- Vasile, B.S.; Chircov, C.; Matei, M.; Neacs, I.A. Iron Oxide—Silica Core—Shell Nanoparticles Functionalized with Essential Oils for Antimicrobial Therapies. Antibiotics 2021, 10, 1138.
- Puiu, R.A.; Balaure, P.C.; Constantinescu, E.; Grumezescu, A.M.; Andronescu, E.; Oprea, O.C.; Vasile, B.S.; Grumezescu, V.; Negut, I.; Nica, I.C.; et al. Anti-cancer nanopowders and maple-fabricated thin coatings based on spions surface modified with paclitaxel loaded β-cyclodextrin. Pharmaceutics 2021, 13, 1356.
- Isela Ruvalcaba Ontiveros, R.; Alberto Duarte Moller, J.; Rocío Carrasco Hernandez, A.; Esperanza Esparza-Ponce, H.; Orrantia Borunda, E.; Deisy Gómez Esparza, C.; Manuel Olivares Ramírez, J. A Simple Way to Produce Gold Nanoshells for Cancer Therapy. In Current Topics in Biochemical Engineering; IntechOpen: London, UK, 2019.
- Khameneh, B.; Diab, R.; Ghazvini, K.; Fazly Bazzaz, B.S. Breakthroughs in bacterial resistance mechanisms and the potential ways to combat them. Microb. Pathog. 2016, 95, 32–42.
- Slavin, Y.N.; Asnis, J.; Häfeli, U.O.; Bach, H. Metal nanoparticles: Understanding the mechanisms behind antibacterial activity. J. Nanobiotechnol. 2017, 15, 65.
- AlMatar, M.; Makky, E.A.; Var, I.; Koksal, F. The Role of Nanoparticles in the Inhibition of Multidrug-Resistant Bacteria and Biofilms. Curr. Drug Deliv. 2018, 15, 470–484.
- Franci, G.; Falanga, A.; Galdiero, S.; Palomba, L.; Rai, M.; Morelli, G.; Galdiero, M. Silver Nanoparticles as Potential Antibacterial Agents. Molecules 2015, 20, 8856–8874.
- Nisar, P.; Ali, N.; Rahman, L.; Ali, M.; Shinwari, Z.K. Antimicrobial activities of biologically synthesized metal nanoparticles: An insight into the mechanism of action. JBIC J. Biol. Inorg. Chem. 2019, 24, 929–941.
- Abdal Dayem, A.; Hossain, M.; Lee, S.; Kim, K.; Saha, S.; Yang, G.-M.; Choi, H.; Cho, S.-G. The Role of Reactive Oxygen Species (ROS) in the Biological Activities of Metallic Nanoparticles. Int. J. Mol. Sci. 2017, 18, 120.
- Snezhkina, A.V.; Kudryavtseva, A.V.; Kardymon, O.L.; Savvateeva, M.V.; Melnikova, N.V.; Krasnov, G.S.; Dmitriev, A.A. ROS Generation and Antioxidant Defense Systems in Normal and Malignant Cells. Oxid. Med. Cell. Longev. 2019, 2019, 6175804.
- Gupta, A.; Mumtaz, S.; Li, C.-H.; Hussain, I.; Rotello, V.M. Combatting antibiotic-resistant bacteria using nanomaterials. Chem. Soc. Rev. 2019, 48, 415–427.
- Ulloa-Ogaz, A.L.; Piñón-Castillo, H.A.; Muñoz-Castellanos, L.N.; Athie-García, M.S.; Ballinas-Casarrubias, M.D.L.; Murillo-Ramirez, J.G.; Flores-Ongay, L.Á.; Duran, R.; Orrantia-Borunda, E. Oxidative damage to Pseudomonas aeruginosa ATCC 27833 and Staphylococcus aureus ATCC 24213 induced by CuO-NPs. Environ. Sci. Pollut. Res. 2017, 24, 22048–22060.




