Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Josep A Rossello | + 1140 word(s) | 1140 | 2021-12-03 03:47:14 | | | |
| 2 | Camila Xu | Meta information modification | 1140 | 2021-12-10 09:44:58 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Rossello, J.A. Interstitial Telomeric-like Repeats (ITR). Encyclopedia. Available online: https://encyclopedia.pub/entry/16887 (accessed on 08 February 2026).
Rossello JA. Interstitial Telomeric-like Repeats (ITR). Encyclopedia. Available at: https://encyclopedia.pub/entry/16887. Accessed February 08, 2026.
Rossello, Josep A. "Interstitial Telomeric-like Repeats (ITR)" Encyclopedia, https://encyclopedia.pub/entry/16887 (accessed February 08, 2026).
Rossello, J.A. (2021, December 08). Interstitial Telomeric-like Repeats (ITR). In Encyclopedia. https://encyclopedia.pub/entry/16887
Rossello, Josep A. "Interstitial Telomeric-like Repeats (ITR)." Encyclopedia. Web. 08 December, 2021.
Copy Citation
Interstitial telomeric repeat (ITR) sites, also known as interstitial telomeric sequences (ITSs), consist of tandem repeats of telomeric motifs that are located within intrachromosomal regions, including repeats located close to the centromeres and the ones found between the centromeres and the telomeres.
interstitial telomeric repeats
in situ hybridisation
chromosomal landmarks
karyological evolution
1. Introduction
The physical package of genetic material is organised in universal structures called chromosomes. In prokaryotes, and in the organelles, chromosomes display a single and circular structure in the absence of a surrounding membrane envelope. However, in the nucleus of most eukaryotes, chromosomes are linear, and their numbers, shape, size, and C-genome size vary greatly among species.
Structurally, a canonical eukaryote chromosome consists basically of chromatids, a centromere, and telomeres, which are preserved during cell division through mitosis and meiosis. Centromeres and telomeres are vital for the integrity of eukaryotic chromosomes. The former play a key role in the precise segregation of chromosomes throughout mitosis and meiosis processes during cell divisions. Meanwhile, telomeres are the terminal DNA-nucleoprotein complexes of chromosomes (Figure 1A), capping their ends and protecting the chromosome against end-to-end fusions [1].
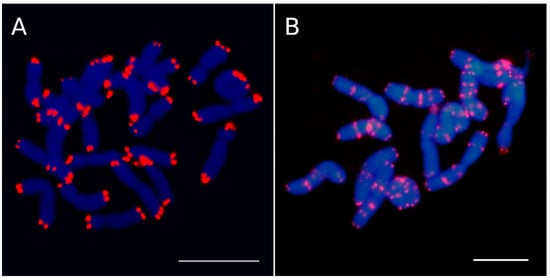
Figure 1. Telomeric sequences (Arabidopsis-type repeats) in (A) Lysimachia minoricensis (Primulaceae) and (B) Anacyclus pyrethrum (Asteraceae). L. minoricensis lacks ITR repeats, whereas A. pyrethrum shows many ITR sites located at proximal and interstitial regions. Scale bars = 10 µm.
Interstitial telomeric repeat (ITR) sites, also known as interstitial telomeric sequences (ITSs), consist of tandem repeats of telomeric motifs that are located within intrachromosomal regions (Figure 1B), including repeats located close to the centromeres and the ones found between the centromeres and the telomeres [2].
Their presence in fungi [3], vertebrates [4][5], and plants [6][7][8][9][10] suggests that (1) the acquisition of telomeric repeats inside chromosomes in unrelated organisms is a convergent event during karyotype evolution, and (2) multiple cytogenetic and molecular mechanisms might have contributed to the diversity of their formation.
2. Molecular Cytogenetic Approaches Used in ITR Detection
In situ hybridisation (ISH) techniques have become one of the most powerful approaches for mapping specific sequences of DNA in plant cytogenetics, including telomere sequences. The basic principles underlying ISH is similar among all the types of experimental variants that have been developed, regardless of whether standard or sophisticated methods were used. However, experimental issues involving the type of the used probes (cloned, synthetic oligonucleotides), probe labeling (nick-translation, PCR-labeling, pre-labeled oligomer), and probe detection (fluorescent, enzymatic) contribute to the sensitivity of ISH approaches [11][12][13][14]. Several technical approaches have been used to date to detect telomeric repeats in plants, including non-Isotopic ISH [15], FISH (fluorescent in situ hybridisation; [6]), PRINS (primer in situ DNA labeling, [16]), PNA-FISH (peptide nucleic acid-FISH, [17]), ND-FISH (non-denaturant-FISH, [18]), PLOPs-FISH (Pre-Labelled Oligomer Probes, [14]), and CO-FISH (chromosome orientation-FISH, [19]). The drawback of molecular cytogenetic methods is that short arrays of telomeric-like sites may be undetectable by ISH [20]. In these cases, DNA sequencing of interstitial chromosomal regions or whole genomes is the best available option [21].
Initially, the location of telomeres in plant chromosomes were identified in Hordeum vulgare and Secale cereale by [15], who also detected interstitial sites. Some years later, a synthetic oligonucleotide (TTTAGGG), representing the canonical Arabidopsis-type repeat was used as a template in PCR and fluorescently labelled to locate telomere repeats in several unrelated flowering plant species [6]. Most of the studies (71.56%) dealing with the cytogenetic mapping of plant telomeres used synthetic oligonucleotide probes for ISH, including the Arabidopsis-type repeat (53.13%; e.g., [7]), the vertebrate-type repeat (TTAGGG) (15.62%; e.g., [22]), other unusual plant-specific telomere sequences (CTCGGTTATGG, TTTTTTAGGG, T4-5AGCA, TTCAGG and TTTCAGG; 2.14%, e.g., [23][24][25][26]), and the Tetrahymena-type repeat (TTGGGG; 0.67%, e.g., [22]), while a significantly lower number (28.44%) used cloned sequences involving the Arabidopsis clone (27.90%; e.g., [27]) or other specific telomeric regions (0.54%; e.g., [17]). The most likely reasons explaining the preferential use of synthetic probes over cloned sequences may be related to the more complex technical requirements and higher costs involved in the handling and conservation of clones. Currently, ITR repeats detected in plants are constituted by the Arabidopsis-type (TTTAGGG), vertebrate-type (TTAGGG), and Cestrum-type (T4-5AGCA) sequences.
3. ITR Sampling in Seed Plants
A total 627 species from 330 genera belonging to 79 families (sensu APG IV) have been karyologically analysed to detect telomeric sequences in seed plants [6][7][8][10][15][28][14][16][17][18][19][22][23][24][25][26][27][29][30][31][32][33][34][35][36][37][38][39][40][41][42][43][44][45][46][47][48][49][50][51][52][53][54][55][56][57][58][59][60][61][62][63][64][65][66][67][68][69][70][71][72][73][74][75][76][77][78][79][80][81][82][83][84][85][86][87][88][89][90][91][92][93][94][95][96][97][98][99][100][101][102][103][104][105][106][107][108][109][110][111][112][113][114][115][116][117][118][119][120][121][122][123][124][125][126][127][128][129][130][131][132][133][134][135][136][137][138][139][140][141][142][143][144][145][146][147][148][149][150][151][152][153][154][155][156][157][158][159][160][161][162][163][164][165][166][167][168][169][170][171][172][173][174][175][176][177][178][179][180][181][182][183][184][185]. These figures sharply contrast with the greater amount of data reported for nuclear ribosomal DNA loci (35S and 5S rDNA families), the most popular chromosomal landmarks used in plant molecular cytogenetics, with data available for 2148 species, 540 genera, and 114 families [186].
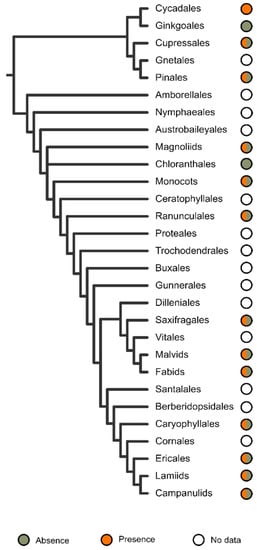
The sampling for detecting telomeric sequences is uneven and biased towards the analysis of large groups, with some exceptions (Figure 2).
Whereas in gymnosperms there is a lack of data only for Gnetales, in angiosperms the number of major groups analysed (11) nearly equals those for which there is no data (13). Speciose ordinal groups not sampled to date are few and include Proteales, Vitales, Santalales, and Cornales, which encompass between 14–151 genera and 590–1750 species [187]. Unfortunately, no species from the three most basal lineages of angiosperms (Amboreallales, Nymphaeales, Austrobaileyales) have been analysed. Although the overall diversity of these orders is fairly limited (12 genera and about 175 species; [187]), their key position in the ancestral diversification of flowering plants makes them priority targets for assessing the presence of ITRs.
4. Taxonomic Distribution of ITRs Is Widespread among Major Lineages of Seed Plants
With the exception of Chloranthales, which exhibits a scarce diversity and has only had three of its analysed, and the monotypic Ginkgoales, all major evolutionary groups sampled had ITRs in their karyotypes (Figure 2 and Figure 3).
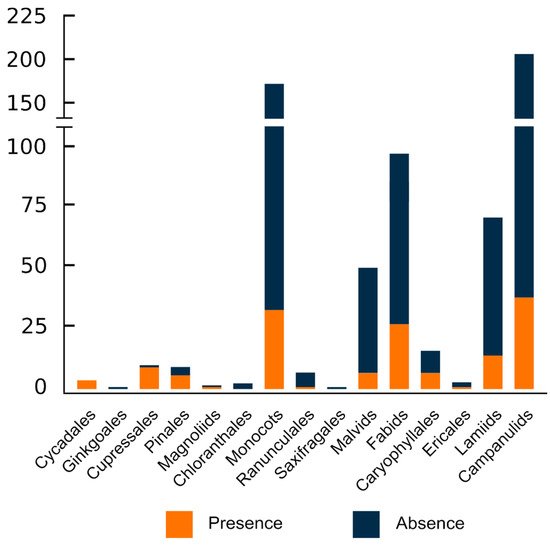
Figure 3. Taxonomic distribution of ITRs in the sampled lineages of seed plants. The number of recorded species is indicated for each group (orange colour). The circumscription of higher taxonomic lineages follows the hypothesis of the Angiosperm Phylogeny Website [187].
However, heterogeneity regarding the distribution at lower taxonomic units is noteworthy. Thus, ITRs occur in only 36 out of 79 sampled families (45.57%), suggesting that disparate results occur within major plant lineages. In gymnosperms, the families with the higher number of ITR occurrences are Podocarpaceae (8 spp.) and Pinaceae (6 spp.), whereas Asteraceae (37 spp.), Fabaceae (21 spp.), and Poaceae (8 spp.) lead among angiosperms. It should be stressed, however, that due to the uneven sampling effort made at different taxonomic levels, these results could be skewed and may not reflect the real values. In this regard, it is worth mentioning that at the family level, the number of species showing ITRs in their karyotypes is strongly correlated to the number of sampled species (Pearson correlation value = 0.994, p < 0.0001).
Overall, lower and similar frequencies of occurrence are attained at the generic and species level. ITRs have been detected in only 88 out of 330 analysed genera (26.67%) and in 149 out of 627 sampled species (23.73%). These results clearly show that although ITRs are widespread in seed plants, their frequency at low taxonomic units is fairly moderate.
References
- Nelson, A.D.; Beilstein, M.A.; Shippen, D.E. Plant telomeres and telomerase. In Molecular Biology; Howell, S.H., Ed.; Springer: New York, NY, USA, 2014; pp. 25–49.
- Lin, K.W.; Yan, J. Endings in the middle: Current knowledge of interstitial telomeric sequences. Mutat. Res. 2008, 658, 95–110.
- Aksenova, A.Y.; Greenwell, P.W.; Dominska, M.; Shishkin, A.A.; Kim, J.C.; Petes, T.D.; Mirkin, S.M. Genome rearrangements caused by interstitial telomeric sequences in yeast. Proc. Natl. Acad. Sci. USA 2013, 110, 19866–19871.
- Ocalewicz, K. Telomeres in fishes. Cytogenet. Genome Res. 2013, 141, 114–125.
- Bolzán, A.D. Interstitial telomeric sequences in vertebrate chromosomes: Origin, function, instability and evolution. Mutat. Res./Rev. Mutat. Res. 2017, 773, 51–65.
- Cox, A.V.; Bennett, S.T.; Parokonny, A.S.; Kenton, A.; Callimassia, M.A.; Bennett, M.D. Comparison of plant telomere locations using a PCR-generated synthetic probe. Ann. Bot. 1993, 72, 239–247.
- Fuchs, J.; Brandes, A.; Schubert, I. Telomere sequence localization and karyotype evolution in higher plants. Plant Syst. Evol. 1995, 196, 227–241.
- Tek, A.; Jiang, J. The centromeric regions of potato chromosomes contain megabase-sized tandem arrays of telomere-similar sequence. Chromosoma 2004, 113, 77–83.
- Gaspin, C.; Rami, J.F.; Lescure, B. Distribution of short interstitial telomere motifs in two plant genomes: Putative origin and function. BMC Plant Biol. 2010, 10, 1–12.
- Mandáková, T.; Gloss, A.D.; Whiteman, N.K.; Lysak, M.A. How diploidization turned a tetraploid into a pseudotriploid. Am. J. Bot. 2016, 103, 1187–1196.
- Menke, M.; Fuchs, J.; Schubert, I. A comparison of sequence resolution on plant chromosomes: PRINS versus FISH. Theor. Appl. Genet. 1998, 97, 1314–1320.
- Jiang, J.; Gill, B.S. Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 2006, 49, 1057–1068.
- Figueroa, D.M.; Bass, H.W. A historical and modern perspective on plant cytogenetics. Brief. Funct. Genom. 2010, 9, 95–102.
- Waminal, N.E.; Pellerin, R.J.; Kim, N.S.; Jayakodi, M.; Park, J.Y.; Yang, T.J.; Kim, H.H. Rapid and efficient FISH using pre-labeled oligomer probes. Sci. Rep. 2018, 8, 1–10.
- Schwarzacher, T.; Heslop-Harrison, J.S. In situ hybridization to plant telomeres using synthetic oligomers. Genome 1991, 34, 317–323.
- Thomas, H.M.; Williams, K.; Harper, J.A. Labelling telomeres of cereals, grasses and clover by primed in situ DNA labelling. Chromosome Res. 1996, 4, 182–184.
- Emadzade, K.; Jang, T.S.; Macas, J.; Kovařík, A.; Novák, P.; Parker, J.; Weiss-Schneeweiss, H. Differential amplification of satellite PaB6 in chromosomally hypervariable Prospero autumnale complex (Hyacinthaceae). Ann. Bot. 2014, 114, 1597–1608.
- Cuadrado, Á.; Golczyk, H.; Jouve, N. A novel, simple and rapid nondenaturing FISH (ND-FISH) technique for the detection of plant telomeres. Potential used and possible target structures detected. Chromosome Res. 2009, 17, 755–762.
- Lou, Q.; Iovene, M.; Spooner, D.M.; Buell, C.R.; Jiang, J. Evolution of chromosome 6 of Solanum species revealed by comparative fluorescence in situ hybridization mapping. Chromosoma 2010, 119, 435–442.
- Majerová, E.; Fojtová, M.; Mandáková, T.; Fajkus, J. Methylation of plant telomeric DNA: What do the results say? Plant Mol. Biol. 2011, 77, 533–536.
- Majerová, E.; Mandáková, T.; Vu, G.T.; Fajkus, J.; Lysak, M.A.; Fojtová, M. Chromatin features of plant telomeric sequences at terminal vs. internal positions. Front. Plant Sci. 2014, 5, 593.
- Sýkorová, E.; Fajkus, J.; Mezníková, M.; Lim, K.Y.; Neplechová, K.; Blattner, F.R.; Leitch, A.R. Minisatellite telomeres occur in the family Alliaceae but are lost in Allium. Am. J. Bot. 2006, 93, 814–823.
- Fajkus, P.; Peška, V.; Sitová, Z.; Fulnečková, J.; Dvořáčková, M.; Gogela, R.; Sýkorová, E.; Fajkus, J. Allium telomeres unmasked: The unusual telomeric sequence (CTCGGTTATGGG)n is synthesized by telomerase. Plant J. 2016, 85, 337–347.
- Peška, V.; Fajkus, P.; Fojtová, M.; Dvořáčková, M.; Hapala, J.; Dvořáček, V.; Polanská, P.; Leitch, A.R.; Sýkorová, E.; Fajkus, J. Characterisation of an unusual telomere motif (TTTTTTAGGG)n in the plant Cestrum elegans (Solanaceae), a species with a large genome. Plant J. 2015, 82, 644–654.
- Sýkorová, E.; Lim, K.Y.; Chase, M.W.; Knapp, S.; Leitch, I.J.; Leitch, A.R.; Fajkus, J. The absence of Arabidopsis-type telomeres in Cestrum and closely related genera Vestia and Sessea (Solanaceae): First evidence from eudicots. Plant J. 2003, 34, 283–291.
- Tran, T.D.; Cao, H.X.; Jovtchev, G.; Neumann, P.; Novák, P.; Fojtová, M.; Vu, G.T.H.; Macas, J.; Fajkus, J.; Schubert, I.; et al. Centromere and telomere sequence alterations reflect the rapid genome evolution within the carnivorous plant genus Genlisea. Plant J. 2015, 84, 1087–1099.
- Katsiotis, A.; Hagidimitriou, M.; Heslop-Harrison, J.S. The close relationship between the A and B genomes in Avena L. (Poaceae) determined by molecular cytogenetic analysis of total genomic, tandemly and dispersed repetitive DNA sequences. Ann. Bot. 1997, 79, 103–109.
- Waminal, N.E.; Pellerin, R.J.; Kang, S.H.; Kim, H.H. Chromosomal mapping of tandem repeats revealed massive chromosomal rearrangements and insights into Senna tora dysploidy. Front. Plant Sci. 2021, 12, 154.
- Hizume, M.; Shibata, F.; Matsusaki, Y.; Kondo, T. Chromosomal localization of telomere sequence repeats in five gymnosperm species. Chromosome Sci. 2000, 4, 39–42.
- Hizume, M.; Kurose, N.; Shibata, F.; Kondo, K. Molecular cytogenetic studies on sex chromosomes and proximal heterochromatin containing telomere-like sequence in Cycas revoluta. Chromosome Sci. 1998, 2, 63–72.
- Shibata, F.; Hizume, M. Survey of Arabidopsis-and human-type telomere repeats in plants using fluorescence in situ hybridisation. Cytologia 2011, 76, 353–360.
- Hizume, M.; Shibata, F.; Matsusaki, Y.; Garajova, Z. Chromosome identification and comparative karyotypic analyses of four Pinus species. Theor. Appl. Genet. 2002, 105, 491–497.
- Shibata, F.; Matsusaki, Y.; Hizume, M. AT-rich sequences containing Arabidopsis-type telomere sequence and their chromosomal distribution in Pinus densiflora. Theor. Appl. Genet. 2005, 110, 1253–1258.
- Schmidt, A.; Doudrick, R.L.; Heslop-Harrison, J.S.; Schmidt, T. The contribution of short repeats of low sequence complexity to large conifer genomes. Theor. Appl. Genet. 2000, 101, 7–14.
- Islam-Faridi, M.N.; Nelson, C.D.; Kubisiak, T.L. Reference karyotype and cytomolecular map for loblolly pine (Pinus taeda L.). Genome 2007, 50, 241–251.
- Murray, B.G.; Friesen, N.; Heslop-Harrison, J.S. Molecular cytogenetic analysis of Podocarpus and comparison with other gymnosperm species. Ann. Bot. 2002, 89, 483–489.
- Kondo, K.; Tagashira, N. Regions in situ-hybridized by the Arabidopsis-type telomere sequence repeats in Zamia chromosomes. Chromosome Sci. 1998, 2, 87–89.
- Kondo, K.; Tagashira, N.; Abd El-Twab, M.H.; Hoshi, Y.; Kokubugata, G.; Honda, Y.; Khaung, K.K. Structural differences of chromosomes in plants detected by fluorescence in situ hybridization using probes of rDNA, Arabidopsis-type telomere sequence repeats and pCrT7-4. In Some Aspects of Chromosome Structure and Functions; Sobti, R.C., Obe, G., Athwal, R.S., Eds.; Springer: Dordrecht, The Netherlands, 2002; pp. 27–35.
- Abd El-Twab, M.H.; Kondo, K. FISH physical mapping of 5S rDNA and telomere sequence repeats identified a peculiar chromosome mapping and mutation in Leucanthemella linearis and Nipponanthemum nipponicum in Chrysanthemum sensu lato. Chromosom. Bot. 2007, 2, 11–17.
- Amosova, A.V.; Bolsheva, N.L.; Samatadze, T.E.; Twardovska, M.O.; Zoshchuk, S.A.; Andreev, I.O.; Badaeva, E.D.; Kunakh, V.A.; Muravenko, O.V. Molecular cytogenetic analysis of Deschampsia antarctica Desv. (Poaceae), Maritime Antarctic. PLoS ONE 2015, 10, e0138878.
- Begum, R.; Alam, S.S.; Menzel, G.; Schmidt, T. Comparative molecular cytogenetics of major repetitive sequence families of three Dendrobium species (Orchidaceae) from Bangladesh. Ann. Bot. 2009, 104, 863–872.
- Begum, R.; Zakrzewski, F.; Menzel, G.; Weber, B.; Alam, S.S.; Schmidt, T. Comparative molecular cytogenetic analyses of a major tandemly repeated DNA family and retrotransposon sequences in cultivated jute Corchorus species (Malvaceae). Ann. Bot. 2013, 112, 123–134.
- Bolsheva, N.L.; Zelenin, A.V.; Nosova, I.V.; Amosova, A.V.; Samatadze, T.E.; Yurkevich, O.Y.; Melnikova, N.V.; Zelenina, D.A.; Volkov, A.A.; Muravenko, O.V. The diversity of karyotypes and genomes within section Syllinum of the genus Linum (Linaceae) revealed by molecular cytogenetic markers and RAPD analysis. PLoS ONE 2015, 10, e0122015.
- Chacón, J.; Sousa, A.; Baeza, C.M.; Renner, S.S. Ribosomal DNA distribution and a genus-wide phylogeny reveal patterns of chromosomal evolution in Alstroemeria (Alstroemeriaceae). Am. J. Bot. 2012, 99, 1501–1512.
- Navrátilová, A.; Neumann, P.; Macas, J. Karyotype analysis of four Vicia species using in situ hybridization with repetitive sequences. Ann. Bot. 2003, 91, 921–926.
- Hanmoto, H.; Kataoka, R.; Ohmido, N.; Yonezawa, Y. Interstitial telomere-like repeats in the Haplopappus gracilis (Asteraceae) genome revealed by fluorescence in situ hybridization. Cytologia 2007, 72, 483–488.
- Cuadrado, Á.; Carmona, A.; Jouve, N. Chromosomal characterization of the three subgenomes in the polyploids of Hordeum murinum L.: New insight into the evolution of this complex. PLoS ONE 2013, 8, e81385.
- Da Silva, C.R.; González-Elizondo, M.S.; Vanzela, A.L. Reduction of chromosome number in Eleocharis subarticulata (Cyperaceae) by multiple translocations. Bot. J. Linn. Soc. 2005, 149, 457–464.
- Deng, H.; Tang, G.; Xu, N.; Gao, Z.; Lin, L.; Liang, D.; Xia, H.; Deng, Q.; Wang, J.; Cai, Z.; et al. Integrated karyotypes of diploid and tetraploid Carrizo Citrange (Citrus sinensis L. Osbeck × Poncirus trifoliata L. Raf.) as determined by sequential multicolor fluorescence in situ hybridization with tandemly repeated DNA sequences. Front Plant Sci. 2020, 11, 569.
- Du, P.; Li, L.N.; Zhang, Z.X.; Liu, H.; Qin, L.; Huang, B.Y.; Dong, W.; Tang, F.; Qi, Z.; Zhang, X.Y. Chromosome painting of telomeric repeats reveals new evidence for genome evolution in peanut. J. Integr. Agric. 2016, 15, 2488–2496.
- Zhang, L.; Yang, X.; Tia, L.; Chen, L.; Yu, W. Identification of peanut (Arachis hypogaea) chromosomes using a fluorescence in situ hybridization system reveals multiple hybridization events during tetraploid peanut formation. New Phytol. 2016, 211, 1424–1439.
- Falistocco, E. Insight into the chromosome structure of the cultivated tetraploid alfalfa (Medicago sativa subsp. sativa L.) by a combined use of GISH and FISH techniques. Plants 2020, 9, 542.
- Dechyeva, D.; Schmidt, T. Molecular organization of terminal repetitive DNA in Beta species. Chromosome Res. 2006, 14, 881–897.
- Zatloukalová, P.; Hřibová, E.; Kubaláková, M.; Suchánková, P.; Šimková, H.; Adoración, C.; Kahl, G.; Millán, T.; Doležel, J. Integration of genetic and physical maps of the chickpea (Cicer arietinum L.) genome using flow-sorted chromosomes. Chromosome Res. 2011, 19, 729–739.
- Gortner, G.; Nenno, M.; Weising, K.; Zink, D.; Nagl, W.; Kahl, G. Chromosomal localization and distribution of simple sequence repeats and the Arabidopsis-type telomere sequence in the genome of Cicer arietinum L. Chromosome Res. 1998, 6, 97–104.
- Grabowska-Joachimiak, A.; Mosiolek, M.; Lech, A.; Góralski, G. C-Banding/DAPI and in situ hybridization reflect karyotype structure and sex chromosome differentiation in Humulus japonicus Siebold & Zucc. Cytogenet. Genome Res. 2011, 132, 203–211.
- Grabowska-Joachimiak, A.; Kula, A.; Gernand-Kliefoth, D.; Joachimiak, A.J. Karyotype structure and chromosome fragility in the grass Phleum echinatum Host. Protoplasma 2015, 252, 301–306.
- He, L.; Liu, J.; Torres, G.A.; Zhang, H.; Jiang, J.; Xie, C. Interstitial telomeric repeats are enriched in the centromeres of chromosomes in Solanum species. Chromosome Res. 2013, 21, 5–13.
- Zhang, Y.; Cheng, C.; Li, J.; Yang, S.; Wang, Y.; Li, Z.; Chen, J.; Lou, Q. Chromosomal structures and repetitive sequences divergence in Cucumis species revealed by comparative cytogenetic mapping. BMC Genom. 2015, 16, 1–13.
- Koo, D.H.; Hong, C.P.; Batley, J.; Chung, Y.S.; Edwards, D.; Bang, J.W.; Hur, Y.; Lim, Y.P. Rapid divergence of repetitive DNAs in Brassica relatives. Genomics 2011, 97, 173–185.
- Kim, E.S.; Bolshev, N.L.; Samatadze, T.E.; Nosov, N.N.; Nosova, I.V.; Zelenin, A.V.; Punina, E.O.; Muravenko, O.V.; Rodionov, A.V. The unique genome of two-chromosome grasses Zingeria and Colpodium, its origin, and evolution. Russ. J. Genet. 2009, 45, 1329–1337.
- Kirov, I.V.; Van Laere, K.; Van Roy, N.; Khrustaleva, L.I. Towards a FISH-based karyotype of Rosa L. (Rosaceae). Comp. Cytogenet. 2016, 10, 543.
- Kondo, K.; Furuta, T. Region in situ-hybridized by the Arabidopsis-type telomere sequence repeats in Drosera chromosomes. Chromosome Sci. 1999, 3, 63–67.
- Kono, Y.; Hoshi, Y.; Setoguchi, H.; Yokota, M.; Oginuma, K. Distribution patterns rDNAs and telomeres and chromosomal rearrangement between two cytotypes of Lysimachia mauritiana L. (Primulaceae). Caryologia 2011, 64, 91–98.
- Kono, Y.; Peng, C.I.; Hoshi, Y.; Yokota, M.; Setoguchi, H.; Lum, S.K.; Oginuma, K. Intraspecific karyotype polymorphism and chromosomal evolution of Lysimachia mauritiana (Primulaceae) in the Ryukyu archipelago of Japan and Taiwan. Cytologia 2019, 84, 93–103.
- Lan, T.Y.; Liu, B.; Dong, F.P.; Chen, R.Y.; Li, X.L.; Chen, C.B. Multicolor FISH analysis of rDNA and telomere on spinach. Front Agric. China 2008, 29, 1405–1408.
- Li, J.; He, S.; Zhang, L.; Hu, Y.; Yang, F.; Ma, L.; Huang, J.; Li, L. Telomere and 45S rDNA sequences are structurally linked on the chromosomes in Chrysanthemum segetum L. Protoplasma 2012, 249, 207–215.
- Luo, X.; Chen, J. Physical map of FISH 5S rDNA and (AG3T3)3 signals displays Chimonanthus campanulatus R.H. Chang & C.S. Ding chromosomes, reproduces its metaphase dynamics and distinguishes its chromosomes. Genes 2019, 10, 904.
- Maravilla, A.J.; Rosato, M.; Álvarez, I.; Nieto Feliner, G.; Rosselló, J.A. Interstitial Arabidopsis-type telomeric repeats in Asteraceae. Plants 2021, submitted.
- Mlinarec, J.; Skuhala, A.; Jurković, A.; Malenica, N.; McCann, J.; Weiss-Schneeweiss, H.; Bohanec, B.; Besendorfer, V. The repetitive DNA composition in the natural pesticide producer Tanacetum cinerariifolium: Interindividual variation of subtelomeric tandem repeats. Front. Plant Sci. 2019, 10, 613.
- Moscone, E.A.; Samuel, R.; Schwarzacher, T.; Schweizer, D.; Pedrosa-Harand, A. Complex rearrangements are involved in Cephalanthera (Orchidaceae) chromosome evolution. Chromosome Res. 2007, 15, 931–943.
- Németh, A.V.; Dudits, D.; Molnár-Láng, M.; Linc, G. Molecular cytogenetic characterisation of Salix viminalis L. using repetitive DNA sequences. J. Appl. Genet. 2013, 54, 265–269.
- Nenno, M.; Zink, D.; Nagl, W. The Arabidopsis telomere sequence is highly abundant in the genome of Phaseolus acutifolius and preferentially located in the centromeres. Rep. Bean Improv. Coop. Nat. Dry Bean Counc. Res. Conf. Ann. Rep. 1998, 41, 103–104.
- Nguyen, T.H.; Waminal, N.E.; Lee, D.S.; Pellerin, R.J.; Ta, T.D.; Campomayor, N.B.; Kang, B.Y.; Kim, H.H. Comparative triple-color FISH mapping in eleven Senna species using rDNA and telomeric repeat probes. Hortic. Environ. Biotechnol. 2021, 62, 927–935.
- Pedrosa, A.; Sandal, N.; Stougaard, J.; Schweizer, D.; Bachmair, A. Chromosomal map of the model legume Lotus japonicus. Genetics 2002, 161, 1661–1672.
- Pellerin, R.J.; Waminal, N.E.; Kim, H.H. Triple-color FISH karyotype analysis of four Korean wild Cucurbitaceae species. Hortic. Sci. Technol. 2018, 36, 98–107.
- Pellerin, R.J.; Waminal, N.E.; Kim, H.H. FISH mapping of rDNA and telomeric repeats in 10 Senna species. Hortic. Environ. Biotechnol. 2019, 60, 253–260.
- Raskina, O.; Barber, J.C.; Nevo, E.; Belyayev, A. Repetitive DNA and chromosomal rearrangements: Speciation-related events in plant genomes. Cytogenet. Genome Res. 2008, 120, 351–357.
- Uchida, W.; Matsunaga, S.; Sugiyama, R.; Kawano, S. Interstitial telomere-like repeats in the Arabidopsis thaliana genome. Genes Genet. Syst. 2002, 77, 63–67.
- Uchida, W.; Matsunaga, S.; Sugiyama, R.; Shibata, F.; Kazama, Y.; Miyazawa, Y.; Hizume, M.; Kawano, S. Distribution of interstitial telomere-like repeats and their adjacent sequences in a dioecious plant, Silene latifolia. Chromosoma 2002, 111, 313–320.
- Rockinger, A.; Sousa, A.; Carvalho, F.A.; Renner, S.S. Chromosome number reduction in the sister clade of Carica papaya with concomitant genome size doubling. Am. J. Bot. 2016, 103, 1082–1088.
- Rosato, M.; Álvarez, I.; Nieto Feliner, G.; Rosselló, J.A. Inter-and intraspecific hypervariability in interstitial telomeric-like repeats (TTTAGGG)n in Anacyclus (Asteraceae). Ann. Bot. 2018, 122, 387–395.
- Sevilleno, S.S.; Ju, Y.H.; Kim, J.S.; Mancia, F.H.; Byeon, E.J.; Cabahug, R.A.; Hwang, Y.J. Cytogenetic analysis of Bienertia sinuspersici Akhani as the first step in genome sequencing. Genes Genom. 2020, 42, 337–345.
- Sousa, A.; Renner, S.S. Interstitial telomere-like repeats in the monocot family Araceae. Bot. J. Linn. Soc. 2015, 177, 15–26.
- Sousa, A.; Cusimano, N.; Renner, S.S. Combining FISH and model-based predictions to understand chromosome evolution in Typhonium (Araceae). Ann. Bot. 2014, 113, 669–680.
- Souza, G.; Vanzela, A.L.; Crosa, O.; Guerra, M. Interstitial telomeric sites and Robertsonian translocations in species of Ipheion and Nothoscordum (Amaryllidaceae). Genetica 2016, 144, 157–166.
- Sýkorová, E.; Lim, K.Y.; Fajkus, J.; Leitch, A.R. The signature of the Cestrum genome suggests an evolutionary response to the loss of (TTTAGGG)n telomeres. Chromosoma 2003, 112, 164–172.
- Vasconcelos, E.V.; Vasconcelos, S.; Ribeiro, T.; Benko-Iseppon, A.M.; Brasileiro-Vidal, A.C. Karyotype heterogeneity in Philodendron s.l. (Araceae) revealed by chromosome mapping of rDNA loci. PLoS ONE 2018, 13, e0207318.
- Waminal, N.E.; Pellerin, R.J.; Jang, W.; Kim, H.H.; Yang, T.J. Characterization of chromosome-specific microsatellite repeats and telomere repeats based on low coverage whole genome sequence reads in Panax ginseng. Plant Breed. Biotechnol. 2018, 6, 74–81.
- Waminal, N.E.; Yang, T.J.; In, J.G.; Kim, H.H. Five-color fluorescence in situ hybridization system for karyotyping of Panax ginseng. Hortic. Environ. Biotechnol. 2020, 61, 869–877.
- Weiss-Schneeweiss, H.; Riha, K.; Jang, C.G.; Puizina, J.; Scherthan, H.; Schweizer, D. Chromosome termini of the monocot plant Othocallis siberica are maintained by telomerase, which specifically synthesises vertebrate-type telomere sequences. Plant J. 2004, 37, 484–493.
- Deng, H.; Xiang, S.; Guo, Q.; Jin, W.; Cai, Z.; Liang, G. Molecular cytogenetic analysis of genome-specific repetitive elements in Citrus clementina Hort. ex Tan. and its taxonomic implications. BMC Plant Biol. 2019, 19, 77.
- Deng, H.; Cai, Z.; Xiang, S.; Guo, Q.; Huang, W.; Liang, G. Karyotype analysis of diploid and spontaneously occurring tetraploid blood orange using multicolor FISH with repetitive DNA sequences as probes. Front. Plant Sci. 2019, 10, 331.
- Prieto, P.; Martín, A.; Cabrera, A. Chromosomal distribution of telomeric and telomeric-associated sequences in Hordeum chilense by in situ hybridization. Hereditas 2004, 141, 122–127.
- Castiglione, M.R.; Kotseruba, V.; Cremonini, R. Methylated-rich regions and tandem repeat arrays along the chromosome complement of Colpodium versicolor (Stev.) Schmalh. Protoplasma 2009, 237, 13.
- Heckmann, S.; Schroeder-Reiter, E.; Kumke, K.; Ma, L.; Nagaki, K.; Murata, M.; Wanner, G.; Houben, A. Holocentric chromosomes of Luzula elegans are characterized by a longitudinal centromere groove, chromosome bending, and a terminal nucleolus organizer region. Cytogenet. Genome Res. 2011, 134, 220–228.
- Maximiano Da Silva, C.M.; González-Elizondo, M.S.; Laforga Vanzela, A.L. Chromosome reduction in Eleocharis maculosa (Cyperaceae). Cytogenet Genome Res. 2008, 122, 175–180.
- Hoshi, Y.; Yagi, K.; Matsuda, M.; Matoba, H.; Tagashira, N.; Pląder, W.; Malepszy, S.; Nagano, K.; Morikawa, A. A comparative study of the three cucumber cultivars using fluorescent staining and fluorescence in situ hybridization. Cytologia 2011, 76, 3–10.
- Pellerin, R.J.; Waminal, N.E.; Belandres, H.R.; Kim, H.H. Karyotypes of three exotic cucurbit species based on triple-color FISH analysis. Korean J. Hortic. Sci. Technol. 2018, 36, 417–425.
- Alexandrov, O.S.; Divashuk, M.G.; Yakovin, N.A.; Karlov, G.I. Sex chromosome differentiation in Humulus japonicus Siebold & Zuccarini, 1846 (Cannabaceae) revealed by fluorescence in situ hybridization of subtelomeric repeat. Comp. Cytogenet. 2012, 6, 239.
- Divashuk, M.G.; Alexandrov, O.S.; Kroupin, P.Y.; Karlov, G.I. Molecular cytogenetic mapping of Humulus lupulus sex chromosomes. Cytogenet Genome Res. 2011, 134, 213–219.
- Hasterok, R.; Ksiazczyk, T.; Wolny, E.; Maluszynska, J. FISH and GISH analysis of Brassica genomes. Acta Biol. Cracov. Bot. 2005, 47, 185–192.
- Adams, S.P.; Hartman, T.P.V.; Lim, K.Y.; Chase, M.W.; Bennett, M.D.; Leitch, I.J.; Leitch, A.R. Loss and recovery of Arabidopsis–type telomere repeat sequences 5′–(TTTAGGG)n–3′ in the evolution of a major radiation of flowering plants. Proc. R. Soc. Lond. B. Biol. Sci. 2001, 268, 1541–1546.
- Jacobs, G.; Dechyeva, D.; Wenke, T.; Weber, B.; Schmidt, T. A BAC library of Beta vulgaris L. for the targeted isolation of centromeric DNA and molecular cytogenetics of Beta species. Genetica 2009, 135, 157–167.
- Puizina, J.; Sviben, T.; Krajačić-Sokol, I.; Zoldoš-Pećnik, V.; Siljak-Yakovlev, S.; Papeš, D.; Besendorfer, V. Cytogenetic and molecular characterization of the Abies alba genome and its relationship with other members of the Pinaceae. Plant Biol. 2008, 10, 256–267.
- Lubaretz, O.; Fuch, J.; Ahne, R.; Meister, A.; Schubert, I. Karyotyping of three Pinaceae species via fluorescent in situ hybridization and computer-aided chromosome analysis. Theor. Appl. Genet. 1996, 92, 411–416.
- de la Herrán, R.; Cuñado, N.; Navajas-Pérez, R.; Santos, J.L.; Rejón, C.R.; Garrido-Ramos, M.A.; Rejón, M.R. The controversial telomeres of lily plants. Cytogenet. Genome Res. 2005, 109, 144–147.
- Sýkorová, E.; Lim, K.Y.; Kunická, Z.; Chase, M.W.; Bennett, M.D.; Fajkus, J.; Leitch, A.R. Telomere variability in the monocotyledonous plant order Asparagales. Proc. R. Soc. Lond. B Biol. Sci. 2003, 270, 1893–1904.
- Monkheang, P.; Chaveerach, A.; Sudmoon, R.; Tanee, T. Karyotypic features including organizations of the 5S, 45S rDNA loci and telomeres of Scadoxus multiflorus (Amaryllidaceae). Comp. Cytogenet. 2016, 10, 637.
- Báez, M.; Souza, G.; Guerra, M. Genome size and cytomolecular diversification in two species of the South African endemic genus Tulbaghia L. (Allioideae, Amaryllidaceae). S. Afr. J. Bot. 2020, 130, 407–413.
- Falistocco, E.; Ferradini, N. Advances in the cytogenetics of Annonaceae, the case of Annona cherimola L. Genome 2020, 63, 357–364.
- Nowicka, A.; Grzebelus, E.; Grzebelus, D. Precise karyotyping of carrot mitotic chromosomes using multicolour-FISH with repetitive DNA. Biol. Plant. 2016, 60, 25–36.
- Zhou, H.C.; Pellerin, R.J.; Waminal, N.E.; Yang, T.J.; Kim, H.H. Pre-labelled oligo probe-FISH karyotype analyses of four Araliaceae species using rDNA and telomeric repeat. Genes Genom. 2019, 41, 839–847.
- Castilho, A.; Vershinin, A.; Heslop-Harrison, J.S. Repetitive DNA and the chromosomes in the genome of oil palm (Elaeis guineensis). Ann. Bot. 2000, 85, 837–844.
- Zaki, N.M. The genome landscape of Elaeis guineensis: Development and Utility of Chromosome-Specific Cytogenetic Markers. Ph.D. Thesis, University of Leicester, Leicester, UK, April 2019.
- Pereira, T.N.S.; Neto, M.F.; de Souza, M.M.; da Costa Geronimo, I.G.; de Melo, C.A.F.; Pereira, M.G. Cytological characterization of Brazilian green dwarf coconut (Cocos nucifera L.) via meiosis and conventional and differential karyotyping. Cytologia 2017, 82, 167–174.
- Robert, M.L.; Lim, K.Y.; Hanson, L.; Sanchez-Teyer, F.; Bennett, M.D.; Leitch, A.R.; Leitch, I.J. Wild and agronomically important Agave species (Asparagaceae) show proportional increases in chromosome number, genome size, and genetic markers with increasing ploidy. Bot. J. Linn. Soc. 2008, 158, 215–222.
- Puizina, J.; Weiss-Schneeweiss, H.; Pedrosa-Harand, A.; Kamenjarin, J.; Trinajstić, I.; Riha, K.; Schweizer, D. Karyotype analysis in Hyacinthella dalmatica (Hyacinthaceae) reveals vertebrate-type telomere repeats at the chromosome ends. Genome 2003, 46, 1070–1076.
- Weiss, H.; Scherthan, H. Aloe spp.–plants with vertebrate-like telomeric sequences. Chromosome Res. 2002, 10, 155–164.
- Abd El-Twab, M.H.; Kondo, K. Physical mapping of 5S, 45S, Arabidopsis-type telomere sequence repeats and AT-rich regions in Achillea millefolium showing intrachromosomal variation by FISH and DAPI. Chromosom. Bot. 2009, 4, 37–45.
- Borgen, L.; Leitch, I.; Santos-Guerra, A. Genome organization in diploid hybrid species of Argyranthemum (Asteraceae) in the Canary Islands. Bot. J. Linn. Soc. 2003, 141, 491–501.
- García, S.; Garnatje, T.; Pellicer, J.; McArthur, E.D.; Siljak-Yakovlev, S.; Vallés, J. Ribosomal DNA, heterochromatin, and correlation with genome size in diploid and polyploid North American endemic sagebrushes (Artemisia, Asteraceae). Genome 2009, 52, 1012–1024.
- Matoba, H.; Uchiyama, H. Physical mapping of 5S rDNA, 18S rDNA and telomere sequences in three species of the genus Artemisia (Asteraceae) with distinct basic chromosome numbers. Cytologia 2009, 74, 115–123.
- Houben, A.; Thompson, N.; Ahne, R.; Leach, C.R.; Verlin, D.; Timmis, J.N. A monophyletic origin of the B chromosomes of Brachycome dichromosomatica (Asteraceae). Plant Syst. Evol. 1999, 219, 127–135.
- Mancia, F.H.; Ju, Y.H.; Lim, K.B.; Kim, J.S.; Nam, S.Y.; Hwang, Y.J. Cytogenetic mapping of Carthamus tinctorius L. with tandemly repeated DNA sequences by fluorescence in situ hybridization. Korean J. Plant Res. 2017, 30, 654–661.
- Dydak, M.; Kolano, B.; Nowak, T.; Siwinska, D.; Maluszynska, J. Cytogenetic studies of three European species of Centaurea L. (Asteraceae). Hereditas 2009, 146, 152–161.
- Cuyacot, A.R.; Won, S.Y.; Park, S.K.; Sohn, S.H.; Lee, J.; Kim, J.S.; Kim, H.H.; Lim, K.M.; Hwang, Y.J. The chromosomal distribution of repetitive DNA sequences in Chrysanthemum boreale revealed a characterization in its genome. Sci. Hortic. 2016, 198, 438–444.
- Abd El-Twab, M.H.; Kondo, K. FISH physical mapping of 5S, 45S and Arabidopsis-type telomere sequence repeats in Chrysanthemum zawadskii showing intra-chromosomal variation and complexity in nature. Chromosom. Bot. 2006, 1, 1–5.
- Cuyacot, A.R.; Lim, K.B.; Kim, H.H.; Hwang, Y.J. Chromosomal characterization based on repetitive DNA distribution in a tetraploid cytotype of Chrysanthemum zawadskii. Hortic. Environ. Biotechnol 2017, 58, 488–494.
- Jamilena, M.; Rejón, C.R.; Rejón, M.R. A molecular analysis of the origin of the Crepis capillaris B chromosome. J. Cell Sci. 1994, 107, 703–708.
- Matoba, H.; Mizutani, T.; Nagano, K.; Hoshi, Y.; Uchiyama, H. Chromosomal study of lettuce and its allied species (Lactuca spp.; Asteraceae) by means of karyotype analysis and fluorescence in situ hybridization. Hereditas 2007, 144, 235–243.
- Pires, J.C.; Lim, K.Y.; Kovarík, A.; Matyásek, R.; Boyd, A.; Leitch, A.R.; Bennett, N.D.; Soltis, P.S.; Soltis, D.E. Molecular cytogenetic analysis of recently evolved Tragopogon (Asteraceae) allopolyploids reveal a karyotype that is additive of the diploid progenitors. Am. J. Bot. 2004, 91, 1022–1035.
- Liu, J.; Luo, X. First report of bicolour FISH of Berberis diaphana and B. soulieana reveals interspecific differences and co-localization of (AGGGTTT)3 and rDNA 5S in B. diaphana. Hereditas 2019, 156, 1–8.
- Mandáková, T.; Marhold, K.; Lysak, M.A. The widespread crucifer species Cardamine flexuosa is an allotetraploid with a conserved subgenomic structure. New Phytol. 2014, 201, 982–992.
- Mandáková, T.; Heenan, P.B.; Lysak, M.A. Island species radiation and karyotypic stasis in Pachycladon allopolyploids. BMC Evol. Biol. 2010, 10, 367.
- Pellerin, R.J.; Waminal, N.E.; Kim, J.Y.; Um, Y.; Kim, H.H. Fluorescence in situ hybridization karyotype analysis of seven Platycodon grandiflorum (Jacq.) A. DC. cultivars. Korean. J. Hortic. Sci. Technol. 2017, 35, 784–792.
- Vanzela, A.L.L.; Cuadrado, Á.; Vieira, A.O.S.; Jouve, N. Genome characterization and relationships between two species of the genus Lobelia (Campanulaceae) determined by repeated DNA sequences. Plant Syst. Evol. 1999, 214, 211–218.
- Divashuk, M.G.; Alexandrov, O.S.; Razumova, O.V.; Kirov, I.V.; Karlov, G.I. Molecular cytogenetic characterization of the dioecious Cannabis sativa with an XY chromosome sex determination system. PLoS ONE 2014, 9, e85118.
- Iovene, M.; Yu, Q.; Ming, R.; Jiang, J. Evidence for emergence of sex-determining gene(s) in a centromeric region in Vasconcellea parviflora. Genetics 2015, 199, 413–421.
- Riha, K.; Fajkus, J.; Siroky, J.; Vyskot, B. Developmental control of telomere lengths and telomerase activity in plants. Plant Cell 1998, 10, 1691–1698.
- Sousa, A.; Fuchs, J.; Renner, S.S. Cytogenetic comparison of heteromorphic and homomorphic sex chromosomes in Coccinia (Cucurbitaceae) points to sex chromosome turnover. Chromosome Res. 2017, 25, 191–200.
- Vanzela, A.L.; Cuadrado, A.; Guerra, M. Localization of 45S rDNA and telomeric sites on holocentric chromosomes of Rhynchospora tenuis Link (Cyperaceae). Genet. Mol. Biol. 2003, 26, 199–201.
- Leitch, A.R.; Lim, K.Y.; Leitch, I.J.; O’Neill, M.; Chye, M.; Low, F. Molecular cytogenetic studies in rubber, Hevea brasiliensis Muell. Arg. (Euphorbiaceae). Genome 1998, 41, 464–467.
- Galasso, I.; Schmidt, T.; Pignone, D. Identification of Lens culinaris ssp. culinaris chromosomes by physical mapping of repetitive DNA sequences. Chromosome Res. 2001, 9, 199–209.
- Hajdera, I.; Siwinska, D.; Hasterok, R.; Maluszynska, J. Molecular cytogenetic analysis of genome structure in Lupinus angustifolius and Lupinus cosentinii. Theor. Appl. Genet. 2003, 107, 988–996.
- Kaczmarek, A.; Naganowska, B.; Wolko, B. Karyotyping of the narrow-leafed lupin (Lupinus angustifolius L.) by using FISH, PRINS and computer measurements of chromosomes. J. Appl. Genet. 2009, 50, 77–82.
- Fonsêca, A.; Ferraz, M.E.; Pedrosa-Harand, A. Speeding up chromosome evolution in Phaseolus: Multiple rearrangements associated with a one-step descending dysploidy. Chromosoma 2016, 125, 413–421.
- Rawlins, D.J.; Highett, M.I.; Shaw, P.J. Localization of telomeres in plant interphase nuclei by in situ hybridization and 3D confocal microscopy. Chromosoma 1991, 100, 424–431.
- Macas, J.; Neumann, P.; Navrátilová, A. Repetitive DNA in the pea (Pisum sativum L.) genome: Comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genom. 2007, 8, 1–16.
- Youn, S.M.; Kim, H.H. Chromosome karyotyping of Senna covesii and S. floribunda based on triple–color FISH mapping of rDNAs and telomeric repeats. Plant Breed. Biotechnol. 2018, 6, 51–56.
- Galasso, I.; Schmidt, T.; Pignone, D.; Heslop-Harrison, J.S. The molecular cytogenetics of Vigna unguiculata (L.) Walp: The physical organization and characterization of 18S-5.8S-25S rRNA genes, 5S rRNA genes, telomere-like sequences, and a family of centromeric repetitive DNA sequences. Theor. Appl. Genet. 1995, 91, 928–935.
- Jankowska, M.; Fuchs, J.; Klocke, E.; Fojtová, M.; Polanská, P.; Fajkus, J.; Schubert, V.; Houben, A. Holokinetic centromeres and efficient telomere healing enable rapid karyotype evolution. Chromosoma 2015, 124, 519–528.
- Xie, S.; Marasek-Ciolakowska, A.; Ramanna, M.S.; Arens, P.; Visser, R.G.; van Tuyl, J.M. Characterization of B chromosomes in Lilium hybrids through GISH and FISH. Plant Syst. Evol. 2014, 300, 1771–1777.
- Lombello, R.A.; Forni-Martins, E.R. Cytogenetics and evolutionary analysis of Lophanthera, an Amazonian arboreal Malpighiaceae. Cytologia 2002, 67, 41–45.
- Islam-Faridi, N.; Sakhanokho, H.F.; Nelson, C.D. New chromosome number and cyto-molecular characterization of the African Baobab (Adansonia digitata L.)—“The Tree of Life”. Sci. Rep. 2020, 10, 1–15.
- Ling, J.; Cheng, H.; Liu, F.; Song, G.L.; Wang, C.Y.; Li, S.H.; Zhang, X.D.; Wang, Y.H.; Wang, K.B. The cloning and fluorescence in situ hybridization analysis of cotton telomere sequence. J. Integr. Agric. 2012, 11, 1417–1423.
- Osuji, J.O.; Crouch, J.; Harrison, G.; Heslop-Harrison, J.S. Molecular cytogenetics of Musa species, cultivars and hybrids: Location of 18S-5.8 S-25S and 5S rDNA and telomere-like sequences. Ann. Bot. 1998, 82, 243–248.
- Luo, X.; Liu, J. Fluorescence in situ hybridization (FISH) analysis of the locations of the oligonucleotides 5S rDNA, (AGGGTTT)3, and (TTG)6 in three genera of Oleaceae and their phylogenetic framework. Genes 2019, 10, 375.
- Golczyk, H.; Massouh, A.; Greiner, S. Translocations of chromosome end-segments and facultative heterochromatin promote meiotic ring formation in evening primroses. Plant Cell 2014, 26, 1280–1293.
- Zhou, H.C.; Park, E.J.; Kim, H.H. Analysis of chromosome composition of Gastrodia elata Blume by fluorescent in situ hybridization using rDNA and telomeric repeat probes. Korean J. Med. Crop Sci. 2018, 26, 113–118.
- de Melo, C.A.F.; Souza, M.M.; Silva, G.S. Karyotype analysis by FISH and GISH techniques on artificial backcrossed interspecific hybrids involving Passiflora sublanceolata (Killip) MacDougal (Passifloraceae). Euphytica 2017, 213, 161.
- Dhar, M.K.; Kaul, S.; Friebe, B.; Gill, B.S. Chromosome identification in Plantago ovata Forsk. through C-banding and FISH. Curr. Sci. 2002, 83, 150–152.
- Shams, I.; Raskina, O. Supernumerary B chromosomes and plant genome changes: A snapshot of wild populations of Aegilops speltoides Tausch (Poaceae, Triticeae). Int. J. Mol. Sci. 2020, 21, 3768.
- Santos, F.C.; Guyot, R.; do Valle, C.B.; Chiari, L.; Techio, V.H.; Heslop-Harrison, P.; Vanzela, A.L.L. Chromosomal distribution and evolution of abundant retrotransposons in plants: Gypsy elements in diploid and polyploid Brachiaria forage grasses. Chromosome Res. 2015, 23, 571–582.
- Rocha, L.C.; de Oliveira Bustamante, F.; Silveira, R.A.D.; Torres, G.A.; Mittelmann, A.; Techio, V.H. Functional repetitive sequences and fragile sites in chromosomes of Lolium perenne L. Protoplasma 2015, 252, 451–460.
- Jiang, J.; Gill, B.S.; Wang, G.L.; Ronald, P.C.; Ward, D.C. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc. Natl. Acad. Sci. USA 1995, 92, 4487–4491.
- Cheng, Z.; Stupar, R.M.; Gu, M.; Jiang, J. A tandemly repeated DNA sequence is associated with both knob-like heterochromatin and a highly decondensed structure in the meiotic pachytene chromosomes of rice. Chromosoma 2001, 110, 24–31.
- Manzanero, S.; Puertas, M.J. Rye terminal neocentromeres: Characterisation of the underlying DNA and chromatin structure. Chromosoma 2003, 111, 408–415.
- Werner, J.E.; Kota, R.S.; Gill, B.S.; Endo, T.R. Distribution of telomeric repeats and their role in the healing of broken chromosome ends in wheat. Genome 1992, 35, 844–848.
- Mlinarec, J.; Papeš, D.A.; Besendorfer, V. Ribosomal, telomeric and heterochromatin sequences localization in the karyotype of Anemone hortensis. Bot. J. Linn. Soc. 2006, 150, 177–186.
- Schuster, M.; Fuchs, J.; Schubert, I. Cytogenetics in fruit breeding-localization of ribosomal RNA genes on chromosomes of apple (Malus × domestica Borkh.). Theor. Appl. Genet. 1997, 94, 322–324.
- Yu, C.; Deng, X.; Chen, C. Chromosomal characterization of a potential model mini-Citrus (Fortunella hindsii). Tree Genet. Genomes 2019, 15, 73.
- Lan, H.; Chen, C.L.; Miao, Y.; Yu, C.X.; Guo, W.W.; Xu, Q.; Deng, X.X. Fragile sites of ‘Valencia’ sweet orange (Citrus sinensis) chromosomes are related with active 45S rDNA. PLoS ONE 2016, 11, e0151512.
- Xin, H.; Zhang, T.; Han, Y.; Wu, Y.; Shi, J.; Xi, M.; Jiang, J. Chromosome painting and comparative physical mapping of the sex chromosomes in Populus tomentosa and Populus deltoides. Chromosoma 2018, 127, 313–321.
- Islam-Faridi, M.N.; Nelson, C.D.; DiFazio, S.P.; Gunter, L.E.; Tuskan, G.A. Cytogenetic analysis of Populus trichocarpa–ribosomal DNA, telomere repeat sequence, and marker-selected BACs. Cytogenet. Genome Res. 2009, 125, 74–80.
- Datson, P.M.; Murray, B.G. Ribosomal DNA locus evolution in Nemesia: Transposition rather than structural rearrangement as the key mechanism? Chromosome Res. 2006, 14, 845–857.
- Zhou, H.C.; Waminal, N.E.; Kim, H.H. In silico mining and FISH mapping of a chromosome-specific satellite DNA in Capsicum annuum L. Genes Genom. 2019, 41, 1001–1006.
- Moscone, E.A.; Scaldaferro, M.A.; Grabiele, M.; Cecchini, N.M.; Sánchez García, Y.; Jarret, R.; Daviña, J.R.; Ducasse, D.A.; Barboza, G.E.; Ehrendorfer, F. The evolution of chili peppers (Capsicum-Solanaceae): A cytogenetic perspective. Acta Hortic. 2007, 745, 137–169.
- Ganal, M.W.; Lapitan, N.L.; Tanksley, S.D. Macrostructure of the tomato telomeres. Plant Cell 1991, 3, 87–94.
- Parokonny, A.S.; Kenton, A.Y.; Gleba, Y.Y.; Bennett, M.D. Genome reorganization in Nicotiana asymmetric somatic hybrids analysed by in situ hybridization. Plant J. 1992, 2, 863–874.
- Peška, V.; Matl, M.; Mandakova, T.; Vitales, D.; Fajkus, P.; Fajkus, J.; García, S. Human-like telomeres in Zostera marina reveal a mode of transition from the plant to the human telomeric sequences. J. Exp. Bot. 2020, 71, 5786–5793.
- Cuadrado, Á.; de Bustos, A.; Jouve, N. On the allopolyploid origin and genome structure of the closely related species Hordeum secalinum and Hordeum capense inferred by molecular karyotyping. Ann. Bot. 2017, 120, 245–255.
- Rocha, L.C.; Mittelmann, A.; Houben, A.; Techio, V.H. Fragile sites of 45S rDNA of Lolium multiflorum are not hotspots for chromosomal breakages induced by X-ray. Mol. Biol. Rep. 2016, 43, 659–665.
- Maluszynska, J.; Heslop-Harrison, J.S. Localization of tandemly repeated DNA sequences in Arabidopsis thaliana. Plant J. 1991, 1, 159–166.
- Fonsêca, A.; Pedrosa-Harand, A. Karyotype stability in the genus Phaseolus evidenced by the comparative mapping of the wild species Phaseolus microcarpus. Genome 2013, 56, 335–343.
- Vitales, D.; D’Ambrosio, U.; Gálvez, F.; Kovařík, A.; García, S. Third release of the plant rDNA database with updated content and information on telomere composition and sequenced plant genomes. Plant Syst. Evol. 2017, 303, 1115–1121.
- Angiosperm Phylogeny Website. Available online: http://www.mobot.org/MOBOT/research/APweb/ (accessed on 9 September 2021).
- Cole, T.C.H.; Bachelier, J.B.; Hilger, H.H. Tracheophyte phylogeny poster—Vascular plants: Systematics and characteristics. Peer J. Prepr. 2019, 7, e2614v3.
- Cole, T.C.H.; Hilger, H.H.; Stevens, P. Angiosperm phylogeny poster (APP)—Flowering plant systematics. Peer J. Prepr. 2019, 7, e2320v6.
More
Information
Subjects:
Genetics & Heredity; Biology; Plant Sciences
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.3K
Entry Collection:
Environmental Sciences
Revisions:
2 times
(View History)
Update Date:
10 Dec 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No