Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Interstitial telomeric repeat (ITR) sites, also known as interstitial telomeric sequences (ITSs), consist of tandem repeats of telomeric motifs that are located within intrachromosomal regions, including repeats located close to the centromeres and the ones found between the centromeres and the telomeres.
- interstitial telomeric repeats
- in situ hybridisation
- chromosomal landmarks
- karyological evolution
1. Introduction
The physical package of genetic material is organised in universal structures called chromosomes. In prokaryotes, and in the organelles, chromosomes display a single and circular structure in the absence of a surrounding membrane envelope. However, in the nucleus of most eukaryotes, chromosomes are linear, and their numbers, shape, size, and C-genome size vary greatly among species.
Structurally, a canonical eukaryote chromosome consists basically of chromatids, a centromere, and telomeres, which are preserved during cell division through mitosis and meiosis. Centromeres and telomeres are vital for the integrity of eukaryotic chromosomes. The former play a key role in the precise segregation of chromosomes throughout mitosis and meiosis processes during cell divisions. Meanwhile, telomeres are the terminal DNA-nucleoprotein complexes of chromosomes (Figure 1A), capping their ends and protecting the chromosome against end-to-end fusions [1].
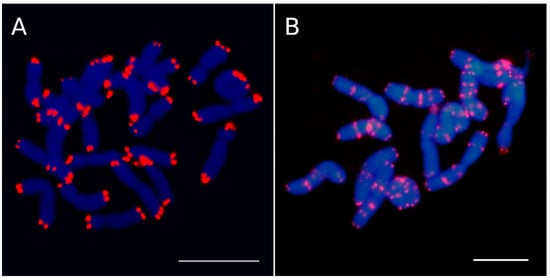
Figure 1. Telomeric sequences (Arabidopsis-type repeats) in (A) Lysimachia minoricensis (Primulaceae) and (B) Anacyclus pyrethrum (Asteraceae). L. minoricensis lacks ITR repeats, whereas A. pyrethrum shows many ITR sites located at proximal and interstitial regions. Scale bars = 10 µm.
Interstitial telomeric repeat (ITR) sites, also known as interstitial telomeric sequences (ITSs), consist of tandem repeats of telomeric motifs that are located within intrachromosomal regions (Figure 1B), including repeats located close to the centromeres and the ones found between the centromeres and the telomeres [2].
Their presence in fungi [3], vertebrates [4,5], and plants [6,7,8,9,10] suggests that (1) the acquisition of telomeric repeats inside chromosomes in unrelated organisms is a convergent event during karyotype evolution, and (2) multiple cytogenetic and molecular mechanisms might have contributed to the diversity of their formation.
2. Molecular Cytogenetic Approaches Used in ITR Detection
In situ hybridisation (ISH) techniques have become one of the most powerful approaches for mapping specific sequences of DNA in plant cytogenetics, including telomere sequences. The basic principles underlying ISH is similar among all the types of experimental variants that have been developed, regardless of whether standard or sophisticated methods were used. However, experimental issues involving the type of the used probes (cloned, synthetic oligonucleotides), probe labeling (nick-translation, PCR-labeling, pre-labeled oligomer), and probe detection (fluorescent, enzymatic) contribute to the sensitivity of ISH approaches [13,14,15,16]. Several technical approaches have been used to date to detect telomeric repeats in plants, including non-Isotopic ISH [11], FISH (fluorescent in situ hybridisation; [6]), PRINS (primer in situ DNA labeling, [17]), PNA-FISH (peptide nucleic acid-FISH, [18]), ND-FISH (non-denaturant-FISH, [19]), PLOPs-FISH (Pre-Labelled Oligomer Probes, [16]), and CO-FISH (chromosome orientation-FISH, [20]). The drawback of molecular cytogenetic methods is that short arrays of telomeric-like sites may be undetectable by ISH [21]. In these cases, DNA sequencing of interstitial chromosomal regions or whole genomes is the best available option [22].
Initially, the location of telomeres in plant chromosomes were identified in Hordeum vulgare and Secale cereale by [11], who also detected interstitial sites. Some years later, a synthetic oligonucleotide (TTTAGGG), representing the canonical Arabidopsis-type repeat was used as a template in PCR and fluorescently labelled to locate telomere repeats in several unrelated flowering plant species [6]. Most of the studies (71.56%) dealing with the cytogenetic mapping of plant telomeres used synthetic oligonucleotide probes for ISH, including the Arabidopsis-type repeat (53.13%; e.g., [7]), the vertebrate-type repeat (TTAGGG) (15.62%; e.g., [23]), other unusual plant-specific telomere sequences (CTCGGTTATGG, TTTTTTAGGG, T4-5AGCA, TTCAGG and TTTCAGG; 2.14%, e.g., [24,25,26,27]), and the Tetrahymena-type repeat (TTGGGG; 0.67%, e.g., [23]), while a significantly lower number (28.44%) used cloned sequences involving the Arabidopsis clone (27.90%; e.g., [28]) or other specific telomeric regions (0.54%; e.g., [18]). The most likely reasons explaining the preferential use of synthetic probes over cloned sequences may be related to the more complex technical requirements and higher costs involved in the handling and conservation of clones. Currently, ITR repeats detected in plants are constituted by the Arabidopsis-type (TTTAGGG), vertebrate-type (TTAGGG), and Cestrum-type (T4-5AGCA) sequences.
3. ITR Sampling in Seed Plants
A total 627 species from 330 genera belonging to 79 families (sensu APG IV) have been karyologically analysed to detect telomeric sequences in seed plants [6,7,8,10,11,12,16,17,18,19,20,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185]. These figures sharply contrast with the greater amount of data reported for nuclear ribosomal DNA loci (35S and 5S rDNA families), the most popular chromosomal landmarks used in plant molecular cytogenetics, with data available for 2148 species, 540 genera, and 114 families [186].
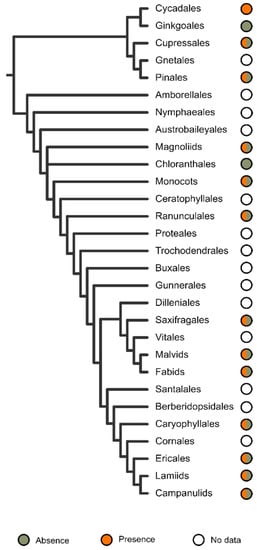
The sampling for detecting telomeric sequences is uneven and biased towards the analysis of large groups, with some exceptions (Figure 2).
Whereas in gymnosperms there is a lack of data only for Gnetales, in angiosperms the number of major groups analysed (11) nearly equals those for which there is no data (13). Speciose ordinal groups not sampled to date are few and include Proteales, Vitales, Santalales, and Cornales, which encompass between 14–151 genera and 590–1750 species [187]. Unfortunately, no species from the three most basal lineages of angiosperms (Amboreallales, Nymphaeales, Austrobaileyales) have been analysed. Although the overall diversity of these orders is fairly limited (12 genera and about 175 species; [187]), their key position in the ancestral diversification of flowering plants makes them priority targets for assessing the presence of ITRs.
4. Taxonomic Distribution of ITRs Is Widespread among Major Lineages of Seed Plants
With the exception of Chloranthales, which exhibits a scarce diversity and has only had three of its analysed, and the monotypic Ginkgoales, all major evolutionary groups sampled had ITRs in their karyotypes (Figure 2 and Figure 3).
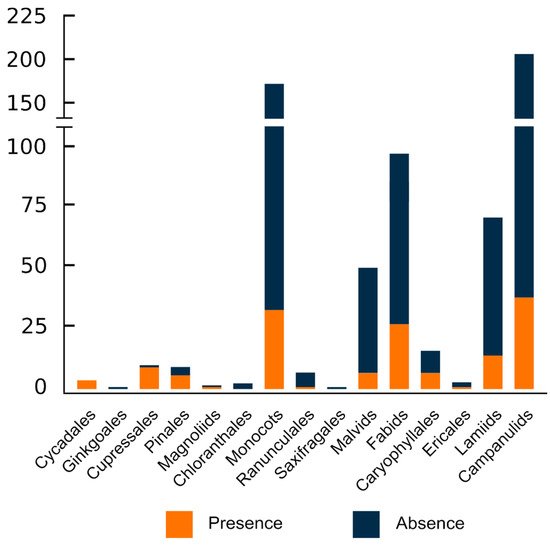
Figure 3. Taxonomic distribution of ITRs in the sampled lineages of seed plants. The number of recorded species is indicated for each group (orange colour). The circumscription of higher taxonomic lineages follows the hypothesis of the Angiosperm Phylogeny Website [187].
However, heterogeneity regarding the distribution at lower taxonomic units is noteworthy. Thus, ITRs occur in only 36 out of 79 sampled families (45.57%), suggesting that disparate results occur within major plant lineages. In gymnosperms, the families with the higher number of ITR occurrences are Podocarpaceae (8 spp.) and Pinaceae (6 spp.), whereas Asteraceae (37 spp.), Fabaceae (21 spp.), and Poaceae (8 spp.) lead among angiosperms. It should be stressed, however, that due to the uneven sampling effort made at different taxonomic levels, these results could be skewed and may not reflect the real values. In this regard, it is worth mentioning that at the family level, the number of species showing ITRs in their karyotypes is strongly correlated to the number of sampled species (Pearson correlation value = 0.994, p < 0.0001).
Overall, lower and similar frequencies of occurrence are attained at the generic and species level. ITRs have been detected in only 88 out of 330 analysed genera (26.67%) and in 149 out of 627 sampled species (23.73%). These results clearly show that although ITRs are widespread in seed plants, their frequency at low taxonomic units is fairly moderate.
This entry is adapted from the peer-reviewed paper 10.3390/plants10112541
This entry is offline, you can click here to edit this entry!