Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Wang, H. LncRNAs Display Circadian Rhythmicity in Zebrafish Larvae. Encyclopedia. Available online: https://encyclopedia.pub/entry/16770 (accessed on 07 February 2026).
Wang H. LncRNAs Display Circadian Rhythmicity in Zebrafish Larvae. Encyclopedia. Available at: https://encyclopedia.pub/entry/16770. Accessed February 07, 2026.
Wang, Han. "LncRNAs Display Circadian Rhythmicity in Zebrafish Larvae" Encyclopedia, https://encyclopedia.pub/entry/16770 (accessed February 07, 2026).
Wang, H. (2021, December 06). LncRNAs Display Circadian Rhythmicity in Zebrafish Larvae. In Encyclopedia. https://encyclopedia.pub/entry/16770
Wang, Han. "LncRNAs Display Circadian Rhythmicity in Zebrafish Larvae." Encyclopedia. Web. 06 December, 2021.
Copy Citation
Long noncoding RNAs (lncRNAs) have been shown to play crucial roles in various life processes, including circadian rhythms. Although next generation sequencing technologies have facilitated faster profiling of lncRNAs, the resulting datasets require sophisticated computational analyses. In particular, the regulatory roles of lncRNAs in circadian clocks are far from being completely understood.
zebrafish larvae
circadian rhythmicity
circadian clocks
noncoding RNAs
lncRNAs
lncRNA-encoded peptides
bioinformatics
1. Introduction
The circadian clock, an endogenous time-keeping mechanism, regulates unique 24-h rhythms of metabolism, physiology and behavior [1]. The suprachiasmatic nucleus (SCN) of the hypothalamus hosts the master clock that drives the circadian rhythms in various tissues and organs [2]. The malfunction of circadian clocks has been closely linked to health problems, such as sleep disorders, mental diseases, and cancers [3]. A variety of model organisms including the fruit fly (Drosophila melanogaster) and the zebrafish (Danio rerio) [4] have been used to study the operating mechanisms of circadian clocks. The fruit fly is an ideal organism to investigate circadian clocks in insects [5] because of its easy genetic manipulation, breeding in a controlled environment, and monitoring of locomotor activities [6]. The zebrafish is also an attractive organism to study the circadian clock in vertebrates [7][8][9]. The ease of obtaining a large number of zebrafish early embryos enables investigation of the onset of circadian rhythmicity [4]. Recently, the genetic dissection of the zebrafish circadian clock has demonstrated that zebrafish share conserved transcription/translation negative feedback loops with fruit flies, mice and humans [10][11][12]. In particular, zebrafish embryos are transparent and do not require feedings for several days post fertilization [13][14]. Hence the confounding effects of feeding can be avoided [15]. As such, zebrafish embryos/larvae allow for studying circadian rhythms independent of feeding.
lncRNAs represent a diverse set of noncoding RNAs that contain more than 200 nucleotides [16]. Interestingly, several lncRNAs have been implicated in regulating circadian rhythms, including circadian rhythms in cancer cells [17][18]. The expression patterns of nearly 100 lncRNAs were shown to be closely linked to the synthesis of hormone melatonin in the rat pineal gland [19]. Melatonin is an integral component of the circadian clock system [7]. The testis is responsible for several key biological functions, such as producing the germ cells and circulating testosterone, the most active androgen [20]. However, the debate over the existence of circadian rhythms in the testis is still unresolved. For example, some studies suggest a lack of circadian clocks in the testis [21], whereas other studies strongly support the presence of circadian activity in the testis [22]. These findings inspired us to investigate the lncRNA-mediated circadian activities in both the pineal gland and testis [8], thereby uncovering 586 and 165 rhythmically expressed lncRNAs in zebrafish pineal gland and testis, respectively. In particular, 26 rhythmically expressed lncRNAs were shown to be coexpressed in both organs [8]. We hypothesize that some lncRNAs are also rhythmically expressed in zebrafish larvae.
Although lncRNAs do not encode canonical proteins, recent studies suggest that they are involved in numerous fundamental biological processes, including determination of cell fate [23], gene regulation [24], transcription, and various diseases [24]. In fact, thousands of lncRNAs have been identified [25] from a diverse set of organisms, including humans [26][27]. For example, GENCODE v7 contains14,880 human lncRNA transcripts [28], while the ZFLNC lncRNA database [29] catalogues over 21,000 zebrafish lncRNAs. Interestingly, numerous lncRNAs [30] have been shown to encode micropeptides, consisting of approximately 100 amino acids [31]. The micropeptides differ from the functional proteins that often contain more than 400 amino acids [32]. The lncRNA-encoded micropeptides have been demonstrated to regulate various biological processes and activities, such as muscle function, transcription, and mRNA stability [33]. Toddler, a lncRNA-encoded microspeptide, regulates Apelin receptors in order to regulate cell movement in zebrafish [34]. A skeletal muscle-specific lncRNA-encoded micropeptide, myoregulin, (MLN) was found to regulate muscle performance [35]. A recent study [36] discovered a conserved 79-amino acid long microprotein, FORCP, encoded by lncRNA LINC00675. A 60-amino acid long micropeptide ASRPS, encoded by lncRNA LINC00908, contained in small open reading frames [37][38]. A micropeptide, miPEP155, encoded by lncRNA MIR155HG was shown to suppress autoimmune inflammation [30]. In particular, our computational analysis recently revealed hundreds of coding lncRNAs in zebrafish [39].
2. LncRNAs Display Circadian Rhythmicity in Zebrafish Larvae
lncRNAs have been implicated in numerous biological processes [24][40]. Although previous studies have uncovered coding potentials [39], expression profiles [26][27][41], and numerous rhythmically expressed lncRNAs from different zebrafish organs [8], our understanding of the involvement of lncRNAs in circadian regulation remains far from complete. Despite a few studies [17][42] investigating the circadian regulation of lncRNAs, the effect of light on rhythmically expressed zebrafish larval lncRNAs has not been studied. Although our previous study [8] identified 26 rhythmically expressed lncRNAs coexpressed in zebrafish pineal gland and testis, further research was needed to investigate how many of these 26 lncRNAs are rhythmically/circadianly expressed in zebrafish larvae.
In this study, we generated time-course transcriptome profiles of zebrafish larvae employing the state-of-the-art bioinformatic tools to investigate circadianly expressed lncRNAs under both DD and LL conditions and uncovered circadian dynamics regulating the expression profiles of the zebrafish larval lncRNAs. In comparison to a recent study [19] that investigated circadian regulation of over one hundred lncRNAs in the rat pineal gland, including elucidation of the operating mechanism of circadian clocks of eight lncRNAs in the suprachiasmatic nucleus (SCN), our study identified thousands of zebrafish larval transcripts under both DD and LL conditions. Specifically, we investigated the expression profiles of 3220 lncRNAs under DD and LL conditions, identified 578 circadianly expressed lncRNAs, and annotated them with GO, COG, and KEGG pathway enrichment analyses. The computational findings suggest that most of these circadianly expressed larval lncRNAs potentially contribute to crucial biological functions.
We compared the circadianly expressed larval lncRNAs with lncRNAs from the pineal gland and testis [8], and found that zebrafish larvae coexpress nine circadianly expressed lncRNAs in both the pineal gland and testis under the DD condition, whereas zebrafish larvae coexpress 12 circadianly expressed lncRNAs with in both the pineal gland and testis under the LL condition, which belong to the 26 lncRNAs coexpressed in zebrafish pineal gland and testis we previously reported [8] (Figure 1A,B). We investigated peptides encoded by these coexpressing lncRNAs to predict their 3D models and functions. In addition, we performed a conservative analysis of the larval lncRNAs with humans, mice, and fruit flies. We found that zebrafish larvae share as many as 35 and 42 lncRNAs with humans under DD and LL conditions, respectively, while zebrafish larvae share as many as one and four lncRNAs with mice under DD and LL conditions, respectively. Hence, we selected the five circadianly expressed lncRNAs shared by these three species, investigated the corresponding lncRNA-encoded peptides, and revealed hundreds of peptides encoded by these 5 lncRNAs. We selected these conserved peptides and investigated their 3D models and corresponding known domains from the Protein Data Bank and uncovered several peptides sharing close resemblance in terms of α-helix, β-strand, and random coils.
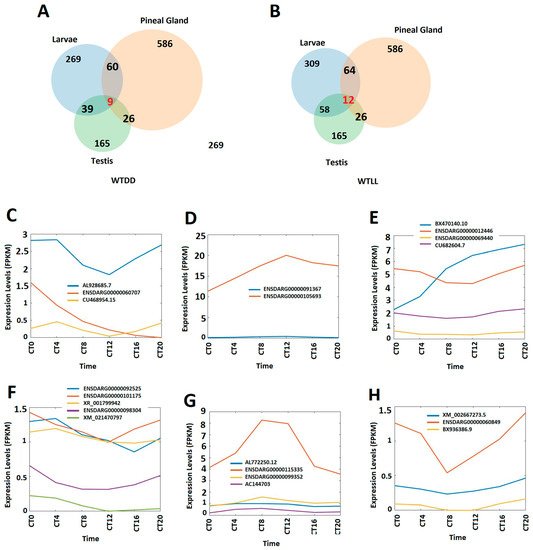
Figure 7. Circadianly expressed zebrafish larval lncRNAs are coexpressed in the zebrafish pineal gland and testis. (A) Circadianly expressed lncRNAs coexpressed in larvae, pineal gland and testis under DD condition. (B) Circadianly expressed lncRNAs coexpressed in larvae, pineal gland and testis under LL condition. (C–E) Expression profiles of representative lncRNAs under DD condition: three zebrafish larval morning (CT0 and CT 4) lncRNAs (C), two zebrafish larval evening (CT8 and CT12) lncRNAs (D), and four zebrafish larval night (CT16 and CT20) lncRNAs (E). (F–H) Expression profiles of representative lncRNAs under LL condition: five zebrafish larval morning lncRNAs (F), four zebrafish larval evening lncRNAs (G), and three zebrafish larval night lncRNAs (H).
Although our framework, which combines novel experimental data with computational analysis, brings unprecedented insights into the circadianly expressed lncRNAs in zebrafish larvae, the study is constrained by a few limitations inherent in the bioinformatic analysis. For example, some of the peptides predicted in this study are more than 100 amino acids long. Hence, additional studies are required to investigate lncRNA-encoded micropeptides that usually contain less than 100 amino acids [31]. However, our approach, which combines biological data and computational techniques, can be applied to investigate both micropeptides and canonical peptides. Second, this study employs RNA-seq technology to investigate the lncRNAs. However, the RNA-seq technology has its own set of shortcomings [43] and often fails to identify certain lncRNAs due to the constraints imposed by poly(A) tails [44]. Third, although our comparative and conservative analysis reveals numerous interesting coexpressing/conserved lncRNAs, the numbers of such lncRNAs are far from complete. It is likely that there are more larval lncRNAs coexpressed in different organs/tissues of zebrafish or conserved with other species. However, due to the lack of experimental data and sequencing information of other tissues, finding a larger number of coexpressing/conserved lncRNAs remains an open research direction. Fourth, the FIMO tool only allows for a few specific p-values to detect the E-Box, D-Box and RORE elements, which may cause multiple false positives. As such, the regulation of circadianly expressed lncRNA by the E-Box, D-Box and RORE requires confirmation by wet-lab experiments. In fact, all the computational predictions require additional biological experimental validation. Fifth, although a zebrafish larva embodies a whole zebrafish, it might not be developed sufficiently to provide the best possible lncRNA expression profiles. A larva needs to undergo a long developmental process before developing an organ such as a testis, and the larval pineal gland and the adult pineal gland may use different sets of lncRNAs. It is possible that some of the lncRNAs expressed in a zebrafish larva may not be expressed in either the adult pineal gland or adult testis. Hence, comparative analysis of zebrafish larval lncRNAs with those in the pineal gland and testis requires additional experimental validations. Sixth, for several larval lncRNAs identified by similarity with ZFLNC lncRNAs, additional research is required to map them to the correct known identifiers in the Gene Bank or Ensembl, as the ZFLNC database lacks identifiers for thousands of lncRNAs. Finally, the effect of light on lncRNAs also requires further investigation. Despite all the limitations, our study uncovers interesting patterns derived from real experimental data. In particular, we predicted 3D models and functions of the conserved peptides encoded by the coexpressing/conserved lncRNAs. To the best of our knowledge, this is for the first time that hundreds of circadianly expressed lncRNAs have been revealed in zebrafish larvae. Our integrative framework, which combines data and bioinformatics analysis, can be expanded to investigate the circadian regulation of a diverse set of noncoding RNAs, and should help circadian biologists to select lncRNAs of interest prior to conducting time-consuming wet-lab experiments.
References
- Young, M.W. Life’s 24-hour clock: Molecular control of circadian rhythms in animal cells. Trends Biochem. Sci. 2000, 25, 601–606.
- Segers, A.; Depoortere, I. Circadian clocks in the digestive system. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 239–251.
- Sancar, A.; Lindsey-Boltz, L.A.; Gaddameedhi, S.; Selby, C.P.; Ye, R.; Chiou, Y.-Y.; Kemp, M.G.; Hu, J.; Lee, J.H.; Ozturk, N. Circadian clock, cancer, and chemotherapy. Biochemistry 2015, 54, 110–123.
- Vatine, G.; Vallone, D.; Gothilf, Y.; Foulkes, N.S. It’s time to swim! Zebrafish and the circadian clock. FEBS Lett. 2011, 585, 1485–1494.
- Beer, K.; Helfrich-Förster, C. Model and Non-model Insects in Chronobiology. Front. Behav. Neurosci. 2020, 14, 221.
- Tataroglu, O.; Emery, P. Studying circadian rhythms in Drosophila melanogaster. Methods 2014, 68, 140–150.
- Ben-Moshe, Z.; Foulkes, N.S.; Gothilf, Y. Functional Development of the Circadian Clock in the Zebrafish Pineal Gland. BioMed Res. Int. 2014, 2014, 235781.
- Mishra, S.K.; Liu, T.; Wang, H. Identification of Rhythmically Expressed LncRNAs in the Zebrafish Pineal Gland and Testis. Int. J. Mol. Sci. 2021, 22, 7810.
- Zhong, Z.; Wang, M.; Huang, G.; Zhang, S.; Wang, H. Molecular Genetic and Genomic Analyses of Zebrafish Circadian Rhythmicity; Springer: New Delhi, India, 2017; pp. 193–209.
- Huang, G.; Zhang, F.; Ye, Q.; Wang, H. The circadian clock regulates autophagy directly through the nuclear hormone receptor Nr1d1/Reverbα and indirectly via Cebpb/(C/ebpβ) in zebrafish. Autophagy 2016, 2, 1292–1309.
- Huang, J.; Zhong, Z.; Wang, M.; Chen, X.; Tan, Y.; Zhang, S.; He, W.; He, X.; Huang, G.; Lu, H.; et al. Circadian Modulation of Dopamine Levels and Dopaminergic Neuron Development Contributes to Attention Deficiency and Hyperactive Behavior. J. Neurosci. 2015, 35, 2572–2587.
- Wang, M.; Zhong, Z.; Zhong, Y.; Zhang, W.; Wang, H. The Zebrafish Period2 Protein Positively Regulates the Circadian Clock through Mediation of Retinoic Acid Receptor (RAR)-related Orphan Receptor α (Rorα). J. Biol. Chem. 2015, 290, 4367–4382.
- Wang, H.; Zhou, Q.; Kesinger, J.W.; Norris, C.; Valdez, C. Heme Regulates Exocrine Peptidase Precursor Genes in Zebrafish. Exp. Biol. Med. 2007, 232, 1170–1180.
- Wang, H.; Kesinger, J.W.; Zhou, Q.; Wren, J.D.; Martin, G.; Turner, S.; Tang, Y.; Frank, M.B.; Centola, M. Identification and characterization of zebrafish ocular formation genes. Genome 2008, 51, 222–235.
- Kelua, J.J.; Pipaliaa, T.G.; Hughes, S.M. Circadian regulation of muscle growth independent of locomotor activity. Proc. Natl. Acad. Sci. USA 2020, 117, 31208–31218.
- The FANTOM Consortium; Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; et al. The Transcriptional Landscape of the Mammalian Genome. Science 2005, 309, 1559–1563.
- Cui, M.; Zheng, M.; Sun, B.; Wang, Y.; Ye, L.; Zhang, X. A Long Noncoding RNA Perturbs the Circadian Rhythm of Hepatoma Cells to Facilitate Hepatocarcinogenesis. Neoplasia 2015, 17, 79–88.
- Fan, Z.; Zhao, M.; Joshi, P.D.; Li, P.; Zhang, Y.; Guo, W.; Xu, Y.; Wang, H.; Zhao, Z.; Yan, J. A class of circadian long non-coding RNAs mark enhancers modulating long-range circadian gene regulation. Nucleic Acids Res. 2017, 45, 5720–5738.
- Coon, S.L.; Munson, P.J.; Cherukuri, P.F.; Sugden, D.; Rath, M.; Møller, M.; Clokie, S.; Fu, C.; Olanich, M.E.; Rangel, Z.; et al. Circadian changes in long noncoding RNAs in the pineal gland. Proc. Natl. Acad. Sci. USA 2012, 109, 13319–13324.
- Bittman, E.L. Timing in the Testis. J. Biol. Rhythm. 2016, 31, 12–36.
- Morse, D.; Cermakian, N.; Brancorsini, S.; Parvinen, M.; Sassone-Corsi, P. No Circadian Rhythms in Testis: Period1 Expression Is Clock Independent and Developmentally Regulated in the Mouse. Mol. Endocrinol. 2003, 17, 141–151.
- Alvarez, J.D.; Sehgal, A. The Thymus Is Similar to the Testis in Its Pattern of Circadian Clock Gene Expression. J. Biol. Rhythm. 2005, 20, 111–121.
- Chen, J.; Wang, Y.; Wang, C.; Hu, J.-F.; Li, W. LncRNA Functions as a New Emerging Epigenetic Factor in Determining the Fate of Stem Cells. Front. Genet. 2020, 11, 277.
- Zhao, T.; Xu, J.; Liu, L.; Bai, J.; Wang, L.; Xiao, Y.; Li, X.; Zhang, L. Computational identification of epigenetically regulated lncRNAs and their associated genes based on integrating genomic data. FEBS Lett. 2015, 589, 521–531.
- Ørom, U.A.; Derrien, T.; Beringer, M.; Gumireddy, K.; Gardini, A.; Bussotti, G.; Lai, F.; Zytnicki, M.; Notredame, C.; Huang, Q.; et al. Long noncoding rnas with enhancer-like function in human cells. Cell 2010, 143, 46–58.
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227.
- Yunusov, D.; Anderson, L.; DaSilva, L.F.; Wysocka, J.; Ezashi, T.R.; Roberts, M.; Verjovski-Almeida, S. HIPSTR and thousands of lncRNAs are heterogeneously expressed in human embryos, primordial germ cells and stable cell lines. Sci. Rep. 2016, 6, 32753.
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Merkel, A.; Gonzalez, D.; Lagarde, J.; et al. The GENCODE v7 Catalogue of Human Long Non-Coding RNAs: Analysis of Their Structure, Evolution and Expression. Genome Res. 2012, 22, 1775–1789.
- Hu, X.; Chen, W.; Li, J.; Huang, S.; Xu, X.; Zhang, X.; Xiang, S.; Liu, C. ZFLNC: A comprehensive and well-annotated database for zebrafish lncRNA. Database 2018, 2018, bay114.
- Niu, L.; Lou, F.; Sun, Y.; Sun, L.; Cai, X.; Liu, Z.; Zhou, H.; Wang, H.; Wang, Z.; Bai, J.; et al. A micropeptide encoded by lncRNA MIR155HG suppresses autoimmune inflammation via modulating antigen presentation. Sci. Adv. 2020, 6, eaaz2059.
- Lin, Y.; Xiao, M.; Chen, H.; Meng, Y.; Zhao, N.; Yang, L.; Tang, H.; Wang, J.; Liu, X.; Zhu, Y.; et al. A novel mitochondrial micropeptide MPM enhances mitochondrial respiratory activity and promotes myogenic differentiation. Cell Death Dis. 2019, 10, 528.
- Sousa, M.E.; Farkas, M.H. Micropeptide. PLoS Genet. 2018, 14, e1007764.
- Xing, J.; Liu, H.; Jiang, W.; Wang, L. LncRNA-Encoded Peptide: Functions and Predicting Methods. Front. Oncol. 2021, 10, 3071.
- Reinier, A.B.; Jaé, N.; Holdt, L.; Dimmeler, S. Long Noncoding RNAs From Clinical Genetics to Therapeutic Targets? J. Am. Coll. Cardiol. 2016, 67, 1214–1226.
- Anderson, D.M.; Anderson, K.M.; Chang, C.-L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A Micropeptide Encoded by a Putative Long Noncoding RNA Regulates Muscle Performance. Cell 2015, 160, 595–606.
- Li, X.L.; Pongor, L.; Tang, W.; Das, S.; Muys, B.R.; Jones, M.F.; Lazar, S.B.; Dangelmaier, A.E.; Hartford, C.C.; Grammatikakis, I.; et al. A small protein encoded by a putative lncRNA regulates apoptosis and tumorigenicity in human colorectal cancer cells. eLife 2020, 9, e53734.
- Wang, Y.; Wu, S.; Zhu, X.; Zhang, L.; Deng, J.; Li, F.; Guo, B.; Zhang, S.; Wu, R.; Zhang, Z.; et al. LncRNA-encoded polypeptide ASRPS inhibits triple-negative breast cancer angiogenesis. J. Exp. Med. 2019, 217, jem.20190950.
- Anfossi, S.; Calin, G.A. When non-coding is not enough. J. Exp. Med. 2020, 217, jem.20192009.
- Mishra, S.K.; Wang, H. Computational Analysis Predicts Hundreds of Coding lncRNAs in Zebrafish. Biology 2021, 10, 371.
- Huang, Y.; Liu, N.; Wang, J.P.; Wang, Y.Q.; Yu, X.L.; Wang, Z.B.; Cheng, X.C.; Zou, Q. Regulatory long non-coding rna and its functions. J. Physiol. Biochem. 2012, 68, 611–618.
- Tovin, A.; Alon, S.; Ben-Moshe, Z.; Mracek, P.; Vatine, G.; Foulkes, N.S.; Jacob-Hirsch, J.; Rechavi, G.; Toyama, R.; Coon, S.L.; et al. Systematic Identification of Rhythmic Genes Reveals camk1gb as a New Element in the Circadian Clockwork. PLoS Genet. 2012, 8, e1003116.
- Ye, M.; Zhang, J.; Wei, M.; Liu, B.; Dong, K. Emerging role of long noncoding RNA-encoded micropeptides in cancer. Cancer Cell Int. 2020, 20, 506.
- Ozsolak, F.; Milos, P.M. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011, 12, 87–98.
- Yu, F.; Zhang, Y.; Cheng, C.; Wang, W.; Zhou, Z.; Rang, W.; Yu, H.; Wei, Y.; Wu, Q.; Zhang, Y. Poly(A)-seq: A method for direct sequencing and analysis of the transcriptomic poly(A)-tails. PLoS ONE 2020, 15, e0234696.
More
Information
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
737
Revisions:
2 times
(View History)
Update Date:
07 Dec 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




