
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Linjing Liu | + 1468 word(s) | 1468 | 2021-08-10 10:47:56 | | | |
| 2 | Catherine Yang | Meta information modification | 1468 | 2021-11-23 03:01:14 | | | | |
| 3 | Catherine Yang | Meta information modification | 1468 | 2021-11-23 03:01:47 | | |
Video Upload Options
Liquid biopsy, which surmounts the limitation of tissue biopsy, is evaluated as a potential tool for early cancer detection and monitoring. By sampling from blood, stool, urine, saliva, and other fluid samples, liquid biopsy provides a non-invasive and feasible cancer detection service. Compared with tissue biopsy, liquid biopsy is also more comprehensive to evaluate tumor heterogeneity since tumor sites can release aberrant signals into body fluid.
1. Introduction
When cells mutate, they could divide uncontrollably and eventually form cancer [1]. According to the World Health Organization, cancer accounts for nearly 10 million deaths in 2020. Unfortunately, this number is estimated to be still climbing in the following decades and will reach 27 million new cases in 2040 [2]. As the second factor of death, cancer accounts for one-sixth of deaths worldwide each year [3]. Therefore, fighting against cancer is a huge challenge for global public health. Early detection, followed by tailored site-specific treatment, plays an important role in the front-line cure of cancer and could reduce the eventual mortality of cancer patients [4][5][6].
Cancer is associated with mutated genes; and genetic analysis is increasingly applied in cancer diagnosis [7]. The traditional methods for genetic testing on cancer patients are sampling from tumor tissues. However, tumor tissue biopsy is limited by several drawbacks such as invasive acquisition, clinical complications, sample preservation, and tumor heterogeneity [8][9][10].
Liquid biopsy [7][11], which surmounts the limitation of tissue biopsy, is evaluated as a potential tool for early cancer detection and monitoring [12]. By sampling from blood, stool, urine, saliva, and other fluid samples, liquid biopsy provides a non-invasive and feasible cancer detection service [13][14][15][16]. Compared with tissue biopsy, liquid biopsy is also more comprehensive to evaluate tumor heterogeneity since tumor sites can release aberrant signals into body fluid [17][18]. Researchers paid significant attention to the different components from liquid biopsy which are associated with cancers [19][20][21][22][23].
As the possibility or severity of tumor in the body is relevant to the liquid biopsy components, accurate cancer prediction based on the characteristics of these components becomes a significant problem. The application of machine learning protocols has been widely studied in recent years, proving to be valuable in early cancer detection. Nevertheless, the required knowledge to implement these methods is high, posing an obstacle to researchers who are looking to get started on liquid biopsy analysis and early cancer detection.
2. Liquid Biopsy Components
During the formation and growth of primary tumors, cells undergo active release, necrosis, or apoptosis [24][25]. In these process, various components are released into the liquid, including circulating tumor cells, cell-free DNA, circulating tumor DNA, cell-free RNA, exosomes, and tumor educated platelets(TEPs) [26].
The presence of circulating Tumor Cells (CTCs) was firstly identified by Ashworth (Australia) in 1869 [27]. When Ashworth performed an autopsy on a metastatic breast cancer patient, cells similar to those from the primary tumor were found in the blood. CTCs are currently defined as the tumor cells that shed or migrate actively into the vessel from the primary tumor or metastatic sites and then circulate in the bloodstream [28]. The opinion of tumor self-seeding suggests that CTCs can recirculate back, resulting in the possibility of metastases, which is responsible for the majority of deaths associated with cancer [29][30]. As the access to peripheral blood circulation is a prerequisite for distant metastasis of tumors [31], detection of tumor cells in blood will indicate the possibility of distant metastasis of tumors [32].
CTC is isolated from peripheral blood, which can avoid invasive and complex biopsy procedures. The culture of tumor cell lines takes a long time and is homogeneous, which cannot accurately reflect the genetic diversity and the changing tumor microenvironment. In contrast, CTCs-derived xenografts can reflect the biological characteristics of cancer more accurately, providing a visual window for studying the dynamic evolution of cancer and allowing monitoring of the longitudinal evolution of tumors at the molecular level.
Platelets (also termed thrombocytes) are the second most abundant cell types in peripheral blood, existing as circulating anucleated cell fragments. The largest platelets are about 2–3 microns in diameter [33]. More recently, platelets are implicated a central role in the local and systemic responses to tumor growth [34][35]. Confrontation of platelets with tumor cells by transferring tumor-associated biomolecules (‘education’) is an emerging research field resulting in the term of tumor-educated platelets (TEPs).
3. Machine Learning Algorithms and Clinical Application in Early Cancer Detection Based on Liquid Biopsy
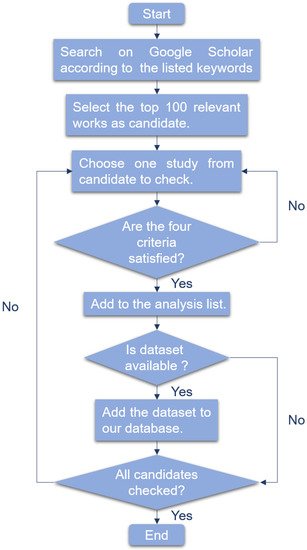
-
The research is about liquid biopsy.
-
The research is about cancer detection.
-
The research utilized corresponding machine learning method.
-
For several models compared, we only consider the model which performs best.
4. Discussion
References
- Nahid, A.A.; Kong, Y. Involvement of machine learning for breast cancer image classification: A survey. Comput. Math. Methods Med. 2017, 2017, 3781951.
- Wild, C.; Weiderpass, E.; Stewart, B. World Cancer Report: Cancer Research for Cancer Prevention; IARC Press: Lyon, France, 2020; pp. 181–188.
- Society, A. Global cancer facts and figures 4th edition. Am. Cancer Soc. 2018, 1, 1–73.
- Cree, I.A.; Uttley, L.; Woods, H.B.; Kikuchi, H.; Reiman, A.; Harnan, S.; Whiteman, B.L.; Philips, S.T.; Messenger, M.; Cox, A.; et al. The evidence base for circulating tumour DNA blood-based biomarkers for the early detection of cancer: A systematic mapping review. BMC Cancer 2017, 17, 697.
- Chen, X.; Gole, J.; Gore, A.; He, Q.; Lu, M.; Min, J.; Yuan, Z.; Yang, X.; Jiang, Y.; Zhang, T.; et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat. Commun. 2020, 11, 1–10.
- WHO. Guide to Cancer Early Diagnosis; WHO: Geneva, Switzerland, 2017.
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472.
- Shinozaki, M.; O’Day, S.J.; Kitago, M.; Amersi, F.; Kuo, C.; Kim, J.; Wang, H.J.; Hoon, D.S. Utility of circulating B-RAF DNA mutation in serum for monitoring melanoma patients receiving biochemotherapy. Clin. Cancer Res. 2007, 13, 2068–2074.
- Zhou, J.; Shi, Y.H.; Fan, J. Circulating cell-free nucleic acids: Promising biomarkers of hepatocellular carcinoma. In Seminars in Oncology; Elsevier: Amsterdam, The Netherlands, 2012; Volume 39, pp. 440–448.
- Cohn, S.L.; Pearson, A.D.; London, W.B.; Monclair, T.; Ambros, P.F.; Brodeur, G.M.; Faldum, A.; Hero, B.; Iehara, T.; Machin, D.; et al. The International Neuroblastoma Risk Group (INRG) classification system: An INRG task force report. J. Clin. Oncol. 2009, 27, 289.
- Diaz Jr, L.A.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579.
- Cai, X.; Janku, F.; Zhan, Q.; Fan, J.B. Accessing genetic information with liquid biopsies. Trends Genet. 2015, 31, 564–575.
- The, L.O. Liquid cancer biopsy: The future of cancer detection? Lancet Oncol. 2016, 17, 123.
- Molina-Vila, M.A.; Mayo-de Las-Casas, C.; Gimenez-Capitan, A.; Jordana-Ariza, N.; Garzón, M.; Balada, A.; Villatoro, S.; Teixido, C.; Garcia-Pelaez, B.; Aguado, C.; et al. Liquid biopsy in non-small cell lung cancer. Front. Med. 2016, 3, 69.
- Strotman, L.N.; Millner, L.M.; Valdes, R.; Linder, M.W. Liquid biopsies in oncology and the current regulatory landscape. Mol. Diagn. Ther. 2016, 20, 429–436.
- Zhang, W.; Chen, X.; Wong, K.C. Noninvasive early diagnosis of intestinal diseases based on artificial intelligence in genomics and microbiome. J. Gastroenterol. Hepatol. 2021, 36, 823–831.
- Chen, M.; Zhao, H. Next-generation sequencing in liquid biopsy: Cancer screening and early detection. Hum. Genom. 2019, 13, 34.
- Peeters, M.; Price, T.; Boedigheimer, M.; Kim, T.W.; Ruff, P.; Gibbs, P.; Thomas, A.; Demonty, G.; Hool, K.; Ang, A. Evaluation of emergent mutations in circulating cell-free DNA and clinical outcomes in patients with metastatic colorectal cancer treated with panitumumab in the ASPECCT study. Clin. Cancer Res. 2019, 25, 1216–1225.
- Cescon, D.W.; Bratman, S.V.; Chan, S.M.; Siu, L.L. Circulating tumor DNA and liquid biopsy in oncology. Nat. Cancer 2020, 1, 276–290.
- Di Meo, A.; Bartlett, J.; Cheng, Y.; Pasic, M.D.; Yousef, G.M. Liquid biopsy: A step forward towards precision medicine in urologic malignancies. Mol. Cancer 2017, 16, 80.
- Heitzer, E.; Perakis, S.; Geigl, J.B.; Speicher, M.R. The potential of liquid biopsies for the early detection of cancer. NPJ Precis. Oncol. 2017, 1, 1–9.
- Ilie, M.; Hofman, V.; Long, E.; Bordone, O.; Selva, E.; Washetine, K.; Marquette, C.H.; Hofman, P. Current challenges for detection of circulating tumor cells and cell-free circulating nucleic acids, and their characterization in non-small cell lung carcinoma patients. What is the best blood substrate for personalized medicine? Ann. Transl. Med. 2014, 2, 107.
- Montani, F.; Marzi, M.J.; Dezi, F.; Dama, E.; Carletti, R.M.; Bonizzi, G.; Bertolotti, R.; Bellomi, M.; Rampinelli, C.; Maisonneuve, P.; et al. miR-Test: A blood test for lung cancer early detection. JNCI J. Natl. Cancer Inst. 2015, 107.
- Gasch, C.; Bauernhofer, T.; Pichler, M.; Langer-Freitag, S.; Reeh, M.; Seifert, A.M.; Mauermann, O.; Izbicki, J.R.; Pantel, K.; Riethdorf, S. Heterogeneity of epidermal growth factor receptor status and mutations of KRAS/PIK3CA in circulating tumor cells of patients with colorectal cancer. Clin. Chem. 2013, 59, 252–260.
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665.
- Alimirzaie, S.; Bagherzadeh, M.; Akbari, M.R. Liquid biopsy in breast cancer: A comprehensive review. Clin. Genet. 2019, 95, 643–660.
- Ashworth, T. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust. Med. J. 1869, 14, 146.
- Imamura, T.; Komatsu, S.; Ichikawa, D.; Kawaguchi, T.; Miyamae, M.; Okajima, W.; Ohashi, T.; Arita, T.; Konishi, H.; Shiozaki, A.; et al. Liquid biopsy in patients with pancreatic cancer: Circulating tumor cells and cell-free nucleic acids. World J. Gastroenterol. 2016, 22, 5627.
- Kim, M.Y.; Oskarsson, T.; Acharyya, S.; Nguyen, D.X.; Zhang, X.H.F.; Norton, L.; Massagué, J. Tumor self-seeding by circulating cancer cells. Cell 2009, 139, 1315–1326.
- Rossi, G.; Mu, Z.; Rademaker, A.W.; Austin, L.K.; Strickland, K.S.; Costa, R.L.B.; Nagy, R.J.; Zagonel, V.; Taxter, T.J.; Behdad, A.; et al. Cell-free DNA and circulating tumor cells: Comprehensive liquid biopsy analysis in advanced breast cancer. Clin. Cancer Res. 2018, 24, 560–568.
- Hayes, D.F.; Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Miller, M.C.; Matera, J.; Allard, W.J.; Doyle, G.V.; Terstappen, L.W. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin. Cancer Res. 2006, 12, 4218–4224.
- Peitzsch, C.; Tyutyunnykova, A.; Pantel, K.; Dubrovska, A. Cancer stem cells: The root of tumor recurrence and metastases. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2017; Volume 44, pp. 10–24.
- Paulus, J.M. Platelet Size in Man; Elsevier: Amsterdam, The Netherlands, 1975.
- Nilsson, R.J.A.; Balaj, L.; Hulleman, E.; Van Rijn, S.; Pegtel, D.M.; Walraven, M.; Widmark, A.; Gerritsen, W.R.; Verheul, H.M.; Vandertop, W.P.; et al. Blood platelets contain tumor-derived RNA biomarkers. Blood J. Am. Soc. Hematol. 2011, 118, 3680–3683.
- Best, M.G.; Sol, N.; Kooi, I.; Tannous, J.; Westerman, B.A.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Koster, J.; et al. RNA-Seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell 2015, 28, 666–676.
- Li, X.; Chen, S.; Hu, X.; Yang, J. Understanding the disharmony between dropout and batch normalization by variance shift. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 2682–2690.
- Loyola-Gonzalez, O. Black-box vs. white-box: Understanding their advantages and weaknesses from a practical point of view. IEEE Access 2019, 7, 154096–154113.
- Lauritsen, S.M.; Kristensen, M.; Olsen, M.V.; Larsen, M.S.; Lauritsen, K.M.; Jørgensen, M.J.; Lange, J.; Thiesson, B. Explainable artificial intelligence model to predict acute critical illness from electronic health records. Nat. Commun. 2020, 11, 1–11.
- Ancona, M.; Ceolini, E.; Öztireli, C.; Gross, M. Towards better understanding of gradient-based attribution methods for deep neural networks. arXiv 2017, arXiv:1711.06104.
- Lundberg, S.; Lee, S.I. A unified approach to interpreting model predictions. arXiv 2017, arXiv:1705.07874.
- Shrikumar, A.; Greenside, P.; Shcherbina, A.; Kundaje, A. Not just a black box: Learning important features through propagating activation differences. arXiv 2016, arXiv:1605.01713.
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should i trust you?” Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144.




