
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Marta Potrykus | + 2138 word(s) | 2138 | 2021-11-03 07:43:40 |
Video Upload Options
The gut microbiota is a crucial factor in maintaining homeostasis. The presence of commensal microorganisms leads to the stimulation of the immune system and its maturation. In turn, dysbiosis with an impaired intestinal barrier leads to accelerated contact of microbiota with the host’s immune cells. Microbial structural parts, i.e., pathogen-associated molecular patterns (PAMPs), such as flagellin (FLG), peptidoglycan (PGN), lipoteichoic acid (LTA), and lipopolysaccharide (LPS), induce inflammation via activation of pattern recognition receptors. Microbial metabolites can also develop chronic low-grade inflammation, which is the cause of many metabolic diseases.
1. Introduction
Berg’s publication was intended to systematize the definition of the microbiome. Based on many definitions, the authors proposed an extended one, in which the microbiome is presented as a combination of microbiota and their theater of activity. Microbiota are living organisms and include bacteria, archaea, fungi, protists, and algae. Viruses, phages, plasmids, viroids, prions, and free DNA or RNA are, by definition, not living organisms, so they are not members of the microbiota, but they belong to the microbiome. The term theater of activity refers to the structural elements of microorganisms, their metabolites, and molecules produced by the host and modified by environmental conditions. The microbiome also includes the given environmental conditions. All of these factors create a complex micro-ecosystem on which the health and well-being of the host largely depend [1].
The gastrointestinal tract is inhabited by more than 10^14 microorganisms, most of which have not been identified [2]. The bacteria are classified into 12 different types, of which 93.5% belong to 4 types: Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria [3]. The gut microbiota plays a major role in digestion, regulation of the immune system, and production of compounds that might alter human metabolism. The microbiota also performs many other functions, i.e., synthesizing vitamins, creating appropriate environmental conditions influencing the oxygen level and pH in the intestines, competing with pathogens (thus reducing their number), and stabilizing the intestinal barrier [4].
The scientific literature shows that the intestinal microbiota can stimulate the immune system and, in the case of excessive stimuli, affect the formation of inflammation.
2. Microbiota-Derived Inflammation
2.1. Pathogen-Associated Molecular Patterns
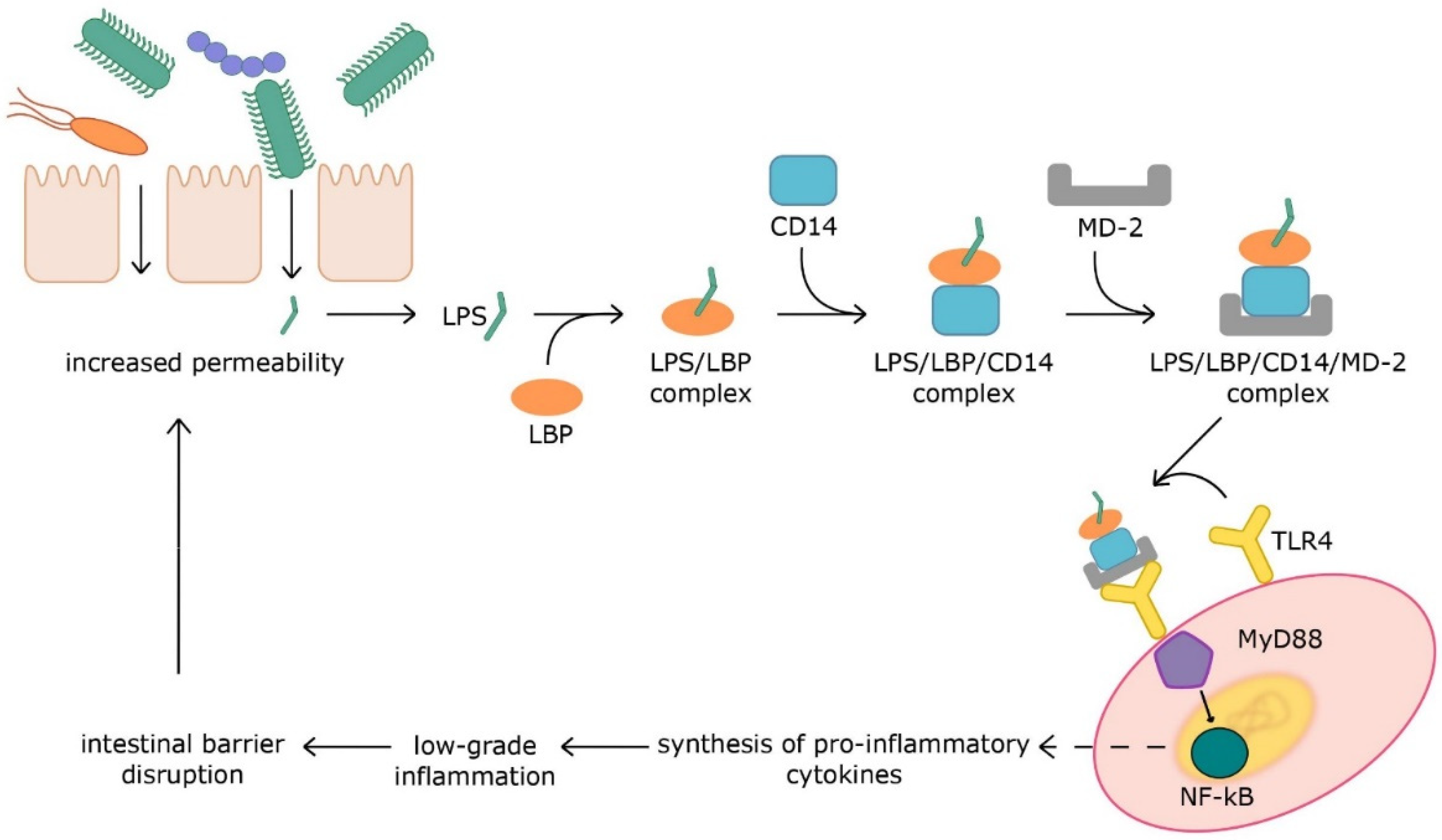
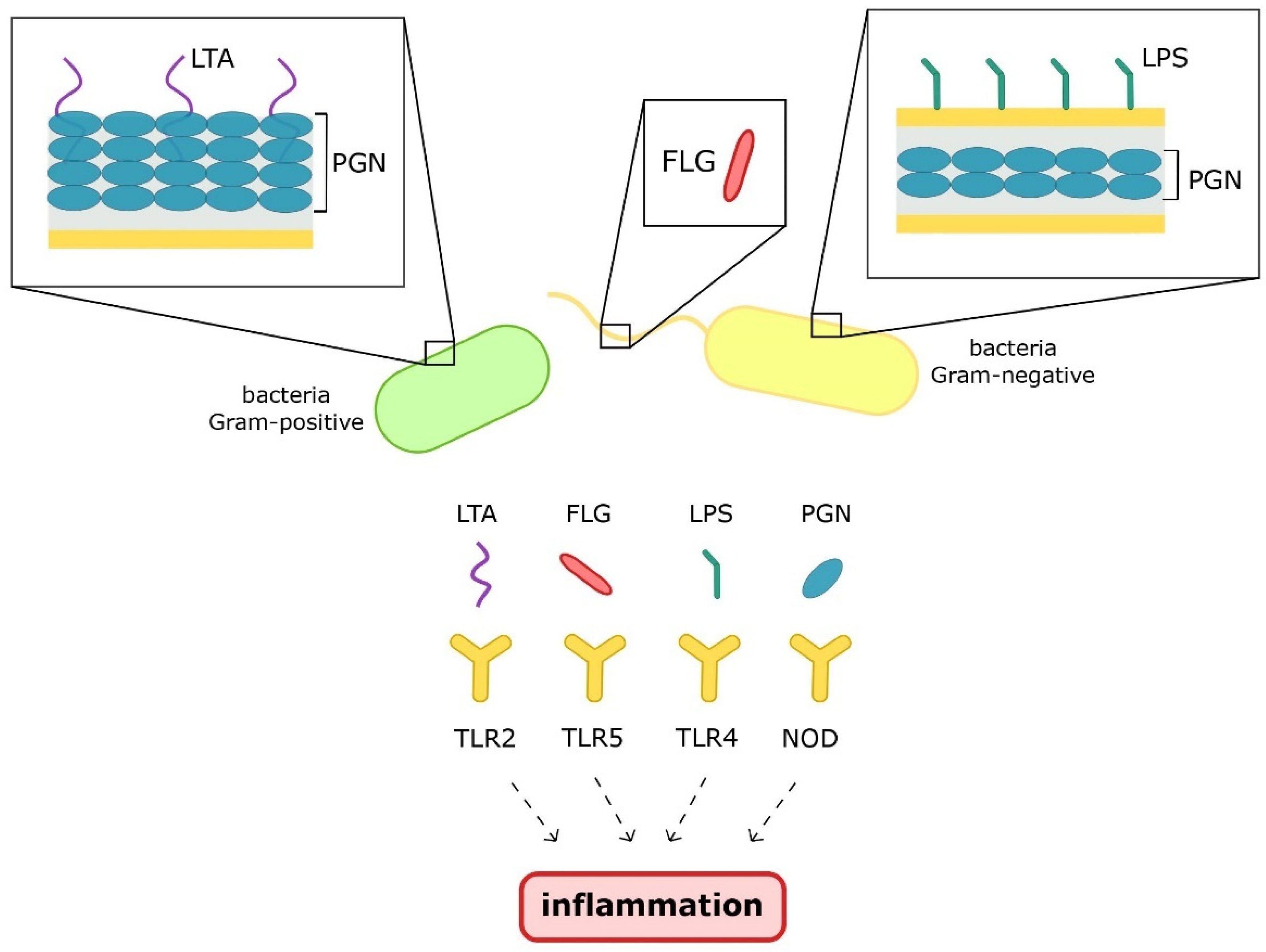
2.2. Metabolites
3. Conclusions
References
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Verges, M.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103.
- Aron-Wisnewsky, J.; Doré, J.; Clement, K. The importance of the gut microbiota after bariatric surgery. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 590–598.
- Lazar, V.; Ditu, L.-M.; Pircalabioru, G.G.; Picu, A.; Petcu, L.; Cucu, N.; Chifiriuc, M.C. Gut Microbiota, host organism, and diet trialogue in diabetes and obesity. Front. Nutr. 2019, 6, 21.
- Scheithauer, T.P.M.; Rampanelli, E.; Nieuwdorp, M.; Vallance, B.A.; Verchere, C.B.; van Raalte, D.H.; Herrema, H. Gut Mi-crobiota as a Trigger for Metabolic Inflammation in Obesity and Type 2 Diabetes. Front. Immunol. 2020, 11, 571731.
- Medzhitov, R. Origin and physiological roles of inflammation. Nature 2008, 454, 428–435.
- Dammermann, W.; Wollenberg, L.; Bentzien, F.; Lohse, A.; Lüth, S. Toll like receptor 2 agonists lipoteichoic acid and pepti-doglycan are able to enhance antigen specific IFNγ release in whole blood during recall antigen responses. J. Immunol. Methods 2013, 396, 107–115.
- Schroder, K.; Tschopp, J. The inflammasomes. Cell 2010, 140, 821–832.
- Rajaee, A.; Barnett, R.; Cheadle, W.G. Pathogen- and danger-associated molecular patterns and the cytokine response in sepsis. Surg. Infect. 2018, 19, 107–116.
- Gnauck, A.; Lentle, R.G.; Kruger, M.C. The Characteristics and Function of Bacterial Lipopolysaccharides and Their Endotoxic Potential in Humans. Int. Rev. Immunol. 2015, 35, 189–218.
- De Santis, S.; Ecavalcanti, E.; Emastronardi, M.; Ejirillo, E.; Echieppa, M. Nutritional keys for intestinal barrier modulation. Front. Immunol. 2015, 6, 612.
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772.
- Mohammad, S.; Thiemermann, C. Role of metabolic endotoxemia in systemic inflammation and potential interventions. Front. Immunol. 2021, 11, 594150.
- Hajam, I.A.; Dar, P.; Shahnawaz, I.; Jaume, J.C.; Lee, J.H. Bacterial flagellin—A potent immunomodulatory agent. Exp. Mol. Med. 2017, 49, e373.
- Yang, J.; Yan, H. TLR5: Beyond the recognition of flagellin. Cell. Mol. Immunol. 2017, 14, 1017–1019.
- Tran, H.Q.; Ley, R.E.; Gewirtz, A.T.; Chassaing, B. Flagellin-elicited adaptive immunity suppresses flagellated microbiota and vaccinates against chronic inflammatory diseases. Nat. Commun. 2019, 10, 1–15.
- Lodes, M.J.; Cong, Y.; Elson, C.O.; Mohamath, R.; Landers, C.J.; Targan, S.R.; Fort, M.; Hershberg, R.M. Bacterial flagellin is a dominant antigen in Crohn disease. J. Clin. Investig. 2004, 113, 1296–1306.
- Radkov, A.D.; Hsu, Y.-P.; Booher, G.; VanNieuwenhze, M.S. Imaging Bacterial Cell Wall Biosynthesis. Annu. Rev. Biochem. 2018, 87, 991–1014.
- Wolf, A.J.; Underhill, D.M. Peptidoglycan recognition by the innate immune system. Nat. Rev. Immunol. 2018, 18, 243–254.
- McDonald, C.; Inohara, N.; Nuñez, G. Peptidoglycan signaling in innate immunity and inflammatory disease. J. Biol. Chem. 2005, 280, 20177–20180.
- Huang, Z.; Wang, J.; Xu, X.; Wang, H.; Qiao, Y.; Chu, W.C.; Xu, S.; Chai, L.; Cottier, F.; Pavelka, N.; et al. Antibody neutral-ization of microbiota-derived circulating peptidoglycan dampens inflammation and ameliorates autoimmunity. Nat. Microbiol. 2019, 4, 766–773.
- Kang, S.-S.; Sim, J.-R.; Yun, C.-H.; Han, S.H. Lipoteichoic acids as a major virulence factor causing inflammatory responses via Toll-like receptor 2. Arch. Pharmacal Res. 2016, 39, 1519–1529.
- Tominari, T.; Sanada, A.; Ichimaru, R.; Matsumoto, C.; Hirata, M.; Itoh, Y.; Numabe, Y.; Miyaura, C.; Inada, M. Gram-positive bacteria cell wall-derived lipoteichoic acid induces inflammatory alveolar bone loss through prostaglandin E production in osteoblasts. Sci. Rep. 2021, 11, 13353.
- Kawahara, K.; Hohjoh, H.; Inazumi, T.; Tsuchiya, S.; Sugimoto, Y. Prostaglandin E2-induced inflammation: Relevance of prostaglandin E receptors. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2014, 1851, 414–421.
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474.
- Yang, S.; Li, X.; Yang, F.; Zhao, R.; Pan, X.; Liang, J.; Tian, L.; Li, X.; Liu, L.; Xing, Y.; et al. Gut microbiota-dependent marker TMAO in Promoting cardiovascular disease: Inflammation Mechanism, clinical prognostic, and potential as a therapeutic target. Front. Pharmacol. 2019, 10, 1360.
- Zhao, S.; Gong, Z.; Zhou, J.; Tian, C.; Gao, Y.; Xu, C.; Chen, Y.; Cai, W.; Wu, J. Deoxycholic Acid Triggers NLRP3 Inflammasome Activation and Aggravates DSS-Induced Colitis in Mice. Front. Immunol. 2016, 7, 536.
- Jia, B.; Jeon, C.O. Promotion and induction of liver cancer by gut microbiome-mediated modulation of bile acids. PLOS Pathog. 2019, 15, e1007954.
- Ramezani, A.; Raj, D.S. The gut microbiome, kidney disease, and targeted interventions. J. Am. Soc. Nephrol. 2014, 25, 657–670.
- Li, D.Y.; Tang, W.W. Contributory Role of Gut Microbiota and Their Metabolites Toward Cardiovascular Complications in Chronic Kidney Disease. Semin. Nephrol. 2018, 38, 193–205.
- Martinez, J.E.; Kahana, D.D.; Ghuman, S.; Wilson, H.P.; Wilson, J.; Kim, S.C.J.; Lagishetty, V.; Jacobs, J.P.; Sinha-Hikim, A.P.; Friedman, T.C. Unhealthy Lifestyle and Gut Dysbiosis: A Better Understanding of the Effects of Poor Diet and Nicotine on the Intestinal Microbiome. Front. Endocrinol. 2021, 12, 667066.
- Gagliardi, A.; Totino, V.; Cacciotti, F.; Iebba, V.; Neroni, B.; Bonfiglio, G.; Trancassini, M.; Passariello, C.; Pantanella, F.; Schippa, S. Rebuilding the gut microbiota ecosystem. Int. J. Environ. Res. Public Health 2018, 15, 1679.




