Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Nataliia Tkachuk | + 3034 word(s) | 3034 | 2021-10-26 08:11:50 | | | |
| 2 | Dean Liu | Meta information modification | 3034 | 2021-11-11 06:38:07 | | | | |
| 3 | Dean Liu | + 1 word(s) | 3035 | 2021-11-12 07:16:35 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Tkachuk, N. Genus Bacillus in the Biodamage/Biodegradation of Plastics. Encyclopedia. Available online: https://encyclopedia.pub/entry/15894 (accessed on 07 February 2026).
Tkachuk N. Genus Bacillus in the Biodamage/Biodegradation of Plastics. Encyclopedia. Available at: https://encyclopedia.pub/entry/15894. Accessed February 07, 2026.
Tkachuk, Nataliia. "Genus Bacillus in the Biodamage/Biodegradation of Plastics" Encyclopedia, https://encyclopedia.pub/entry/15894 (accessed February 07, 2026).
Tkachuk, N. (2021, November 11). Genus Bacillus in the Biodamage/Biodegradation of Plastics. In Encyclopedia. https://encyclopedia.pub/entry/15894
Tkachuk, Nataliia. "Genus Bacillus in the Biodamage/Biodegradation of Plastics." Encyclopedia. Web. 11 November, 2021.
Copy Citation
Bacteria of the genus Bacillus are able to form corrosive compounds, polymer-degrading compounds, antimicrobials and antibiofilm-forming compounds.
Bacillus
biodamage of materials
biodegradation of materials
1. Introduction
In the context of rapid development of technology and construction, as well as creation of new materials, the issue of material biodamage and appropriate protection appears as the focus of substantial research [1][2][3]. Biodamage (biodegradation) of materials can be responsible for human-made ecological catastrophes [1]. Bactericides have traditionally been used to prevent microbiologically influenced corrosion (MIC), although they pose health, safety and environmental problems [4]. Derivatives of substandard pesticides are proposed as inhibitors of MIC [5].
On the other hand, the processes of biodegradation involve chemical elements in geochemical cycles leading to the removal of xenobiotics from the environment [3].
As an alternative, the use of microorganisms is suggested as a protective means against biodamage [6]. However, the research in the field of biological control has been conducted under laboratory conditions and its effectiveness has not yet been confirmed [7].
Sulfate-reducing bacteria (SRB) are most commonly associated with MIC, while little attention has thus far been paid to the representatives of other taxonomic groups [7].
2. Bacteria of the Genus Bacillus in the Biodegradation of Some Synthetic Plastics
Bacteria of the genus Bacillus biodegrade polyethylene (PE), polypropylene (PP), polystyrene (PS), polyvinyl chloride (PVC), poly (ethylene terephthalate) (PET) and polyurethane (PUR). These polymers are synthesized from fossil hydrocarbons, a non-renewable resource, and comprise more than 90% of the global total production of polymeric materials [8]. Enzymes released by microorganisms during growth and development act as aggressive media in biodamage of insulating materials [1]. In particular, such compounds are able to produce the bacterium B. mesentericus (B. subtilis), found on polymer coatings [1]. Degradation of polyethylene can be classified as abiotic (caused by temperature and UV irradiation) or biotic (biodegradation caused by the action of microorganisms). Microorganisms modify and consume the polymer, leading to changes in its properties. Bacteria of the genus Bacillus, among other bacterial strains, are involved in the biodegradation of polyethylene [9][10]. B. amyloliquefaciens, B. brevis, B. cereus, B. circulans, B. halodenitrificans, B. mycoides, B. pumilus, B. sphericus and B. thuringiensis appear to be particularly active agents causing biodegradation of this material. Restrepo-Flórez et al. [9] highlight that although the damage to polyethylene is connected to only one of these two damage modes, both typically act cooperatively in nature [11]. The abiotic mechanisms of deterioration of polyethylene have been described in [11]. The biodegradation of polyethylene and mechanisms associated with this process are analyzed in review publications [9][10]. The biodegradation of polyethylene under normal conditions is extremely slow, while the knowledge of complete metabolic pathways involved in the process, as well as the inventory and structure of respective enzymes, is quite basic [9]. The authors discuss changes of PE in the presence of microorganisms: (1) functional groups on the surface; (2) the surface topography; (3) the hydrophobicity/hydrophilicity of a surface; (4) mechanical properties; (5) crystallinity; (6) molecular weight distribution. It is indicated that the loss of molecular weight and the oxidation of the molecules are the result of synergistic effects caused by biotic and abiotic factors (photo-oxidation or heat treatment) [9]. In particular, thermo-irradiation pretreatment was important for the biodegradation of PE by Bacillus amyloliquefaciens (B. amyloliquefaciens) [12]. Biofilm colonization is the initial step in the degradation of this polymer [9]. Bacteria that form biofilms are capable of producing surfactants, molecules that can mediate the attachment process of microorganisms to a hydrophobic surface [9]. Hydrocarbon polymers are resistant to biodegradation, which results from highly stable C-C and C-H covalent bonds [13]. Restrepo-Flórez et al. [9] note the theoretical possibility of polyethylene being used as a carbon source for microorganisms, similar to many other hydrocarbons, but subject to the reduction in PE molecular weight by the chemical/biochemical processes. The presence, in some members of the genus Bacillus, of enzymes related to the Pseudomonas putida GPo1 membrane-bound alkane hydroxylase system [14] increases this probability. Such enzymes are involved in a complicated chemical reaction: the regio- and stereospecific activation of C-H bonds. There are reports that the weight loss of degradable polyethylene (PE with chemical or photoinitiators or both) positively correlated with CO2 emission in the consortium of Pseudomonas frequentans + B. mycoides under the condition of treatments with or without preheating [15]. Based on this, the researchers conclude that the disposal of degradable polyethylene by these bacteria is a source of carbon.
The initial step involves hydroxylation of C-C bonds, generating primary or secondary alcohols, which are further oxidized to aldehydes or ketones, and then to carboxylic acids [16]. Thus, microbial oxidation decreases the number of carbonyl groups due to the formation of carboxylic acids. Carboxylated n-alkanes are analogous to fatty acids, which can be catabolized by bacteria via the β-oxidation system pathway (Figure 1). However, neither the breaking of C-C bonds within the backbone of PE polymers nor the generation of long carbon chain carboxylic acid hydrolysis products have been reported [16].
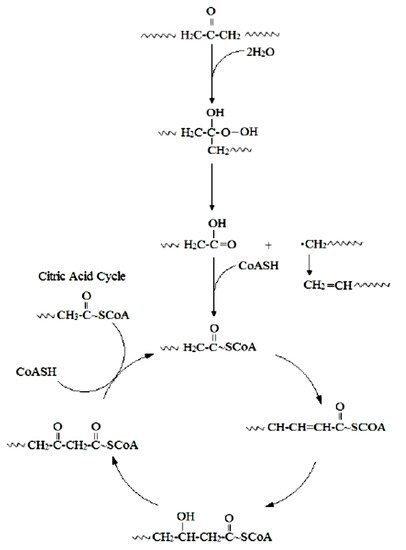
Figure 1. Mechanism for the biodegradation of PE [16].
Thus, bacterial isolates B. amyloliquefaciens contributed to the depolymerization of biodegraded products in extracellular media, indicating the biodegradation process of PE [17]. The participation of B. brevis in PE biodegradation is noted in the work of Wasserbauer et al. [18]. B. cereus degraded UV-irradiated PE associated with a pronounced extracellular production of both laccases and MnP [19]. Biodegradable properties against PE were also observed for B. cereus strain Ma-Su CECRI-1 [20] and B. cereus strain A5,a [21]. In the latter case, a high biodegradable activity (35.72 ± 4.01%) in relation to PE was registered [21]. The ability to biodegrade PE is also inherent to B. subtilis [22]. B. subtilis produced surface active compounds (biosurfactants) that enhance the degradation process of PE [23].
The ability of B. mycoides and B. subtilis (Bacillus species indigenous to the Niger Delta mangrove soil) to biodegrade polyethylene was studied by Ibiene et al. [24].
Bacillus safensis and B. amyloliquefaciens have demonstrated the biodegradable ability for low-density polyethylene thermoplastic. Moreover, B. safensis had better biodegradable ability than B. amyloliquefaciens, shown by the registered weight reduction [25].
It is worth noting that only genus (Bacillus) identification of bacteria has been established in most studies. Thus, in a number of studies, Bacillus sp. are indicated as polyethylene-degrading bacteria [26][27][28][29][30][31], the ability of which to form a biofilm on the surface of this material and reduce its hydrophobicity by isolated bacteria was noted in the article of Yang et al. [29]. The biofilm formation on the surface of polyethylene and its biodegradation were also observed for B. mycoides [15].
Polypropylene was biodegraded by the B. flexus + Pseudomonas azotoformans and B. flexus + B. subtilis associations [32]. Polypropylene was subjected to biodegradation by B. cereus [33][34], B. thuringenesis and B. licheniformis [34]. The latter three species are also involved in the biodegradation of polypropylene-poly-L-lactide [34]. Degradation of PP microplastics by B. gottheilii caused a weight loss of 3.6% after 40 days [35].
Biodegradation of polypropylene has been improved by using polymer blends of carbohydrates, starch or cellulose blends such as that reported for polyethylene and polystyrene [36]. The use of the blends facilitates adhesion of the microorganisms to the surface of the polymer and acts as a co-metabolite. Biodegradation of polycaprolactone blended PP has also been demonstrated using lipase since lipase is well known to degrade the ester linkages of polycaprolactone [36].
Bacillus sp. degrades polystyrene [37][38][39]. The following studies provide insights into biodegradation of PS caused by B. aryabhattai [40], B. subtilis [41], and B. paralicheniformis [42]. B. cereus and B. gottheilii reduced PS granule weight by 7.4% and 5.8%, respectively, within 40 days [35]. B. cereus was also noted as a biodegradable PS in [43].
Strains B. subtilis and B. cereus formed biofilm on PET and poly (lactic acid), demonstrating biochemical activity and accelerating biodegradation of both plastic materials [44]. B. cereus was also identified as part of two consortia capable of biofilm formation and PET degradation [45]. B. megaterium formed a biofilm on PET film [8]. Carboxylesterases from B. licheniformis and B. subtilis partially hydrolyzed PET fibers and showed a high activity against PET oligomers [46].
The mechanism of PET biodegradation is demonstrated in Figure 2. The PET-digesting enzyme labeled as PETase converts PET to mono(2-hydroxyethyl) terephthalic acid (MHET), with minimal amounts of terephthalic acid (TPA) and bis(2-hydroxyethyl)-TPA as secondary products [36].
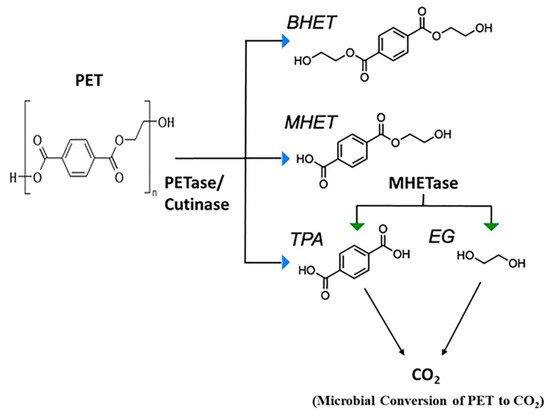
Figure 2. Microbial degradation of PET [36]. PETase, polyethylene terephthalate (PET) hydrolase or PET-digesting enzyme; BHET, bis(2-hydroxyethyl) terephthalic acid; MHET, mono(2-hydroxyethyl) terephthalic acid; TPA, terephthalic acid; EG, ethylene glycol.
B. flexus has been proved to biodegrade poly (vinyl chloride) (PVC) film [47]. Bacteria of this species have been able to form dense biofilm on the plastic film surface and cause a decrease in the mean molecular weight of the PVC film [47]. Bacillus sp., triggering degradation of PVC, caused weight loss of the material of 0.26 ± 0.02% in comparison to initial weights after 90 days [31].
The involvement of bacteria of the genus Bacillus in the biodegradation of polyurethanes is discussed in a review article by G.T. Howard [48]. PUR-degrading bacteria are B. chitinolyticus and B. pumilus [49] and B. subtilis [50][51][52].
Oxidative degradation has been generally associated with poly(ether-urethane) [53]. This mechanism is illustrated by the following reactions.
Thus, the reactive oxygen species initiate degradation of polyether urethanes through oxidative attack of the soft segment. This reactive oxygen species abstracts an alpha-methylene hydrogen atom from the polyether soft segment. The addition of a hydroxyl radical to the carbon radical forms a hemiacetal, which oxidizes to an ester. Acid hydrolysis of the ester results in chain scission of the soft segment and formation of acid end groups. Significant chain scission results in the solubilization and extraction of low molecular weight degradation products [53].
A similar oxidative mechanism was proposed for hard segment degradation [53]. Oxygen radicals abstract an alpha-methylene hydrogen atom from the chain extender at the urethane. Additional hydroxyl radicals combine with the chain radical to form a highly reactive carbonyl hemiacetal. Oxidative hydrolysis of the carbonyl hemiacetal results in chain scission and formation of an unstable carbamic acid and carboxylic acid end groups [53]. The carbamic acid decarboxylates readily to form a free amine. The mechanism of polyurethane biodegradation with the participation of polyurethane esterases is shown by Mohanan et al. [36] (Figure 3).
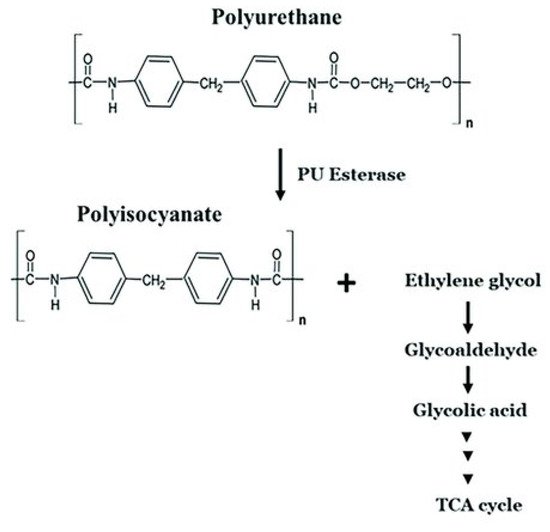
Figure 3. Microbial degradation pathway of polyurethane [36]. TCA cycle: the cycle of tricarboxylic acids. PU esterase: polyurethane esterase.
Ru et al. [54] present the metabolic pathways of degradation of the plastics considered by us (Figure 4).
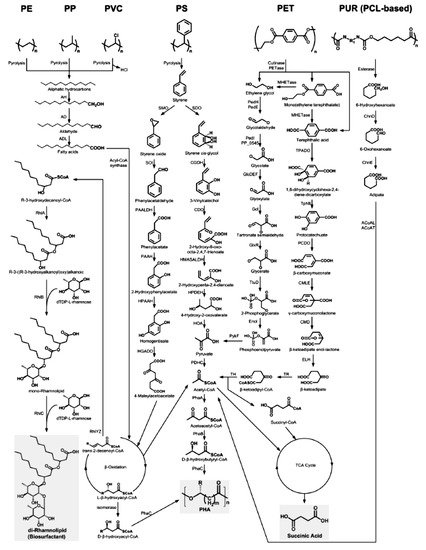
Figure 4. The metabolic pathways of depolymerization products of six kinds of plastics [54]. Plastics: PE, polyethylene; PS, polystyrene; PP, polypropylene; PVC, polyvinyl chloride; PUR, polyurethane; PCL, polycaprolactone diol; PET, polyethylene terephthalate. Enzymes: AH, alkane hydroxylase; AD, alcohol dehydrogenase; ALD, aldehyde dehydrogenase; RhlYZ, R-specific enoyl-CoA hydratase; RhlA, HAA synthetase; RhlB, rhamnosyltransferase 1; RhlC, rhamnosyltransferase 2; SMO, styrene monooxgenase; SOI styrene oxide isomerase; PAALDH, phenacetaldehyde dehydrogenase; PAAH, phenylacetate hydroxylase; HPAAH, 2-hydroxyphenylacetate hydroxylase; HGADO, homogentisate 1,2-dioxygenase; SDO, styrene dioxygenase; CGDH, cis-glycol dehydrogenase; CDO, catechol 2,3-dioxygenase; HMASALDH, 2-hydroxymuconic acid semialdehyde hydrolase; HPDEH, 2-hydroxypenta-2,4-dienoate hydratase; HOA, 4-hydroxy-2-oxovalerate aldolase; PDHC, pyruvate dehydrogenase complex; PhaA, b-ketothiolase; PhaB acetoacetyl-CoA reductase; PhaC, PHA synthase; PedH, quinoprotein alcohol dehydrogenase; PedE, quinoprotein alcohol dehydrogenase; PedI, aldehyde dehydrogenase family protein; PP_0545, aldehyde dehydrogenase family protein; GlcDEF, glycolate oxidase; Gcl glyoxylate carboligase; GlxR, tartronate semialdehyde reductase; TtuD, hydroxypyruvate reductase; PykF, pyruvate kinase; TPADO, TPA dioxygenase; TphB, 1,2-dihydroxy-3,5-cyclohexadiene-1,4-dicarboxylate dehydrogenase; PCDO, protocatechuate 3,4-dioxygenase; CMLE, b-carboxy-cis,cis-muconate lactonizing enzyme; CMD, b-carboxymuconolactone decarboxylase; ELH, enollactone hydrolase; TR, b-ketoadipate:succinyl-CoA transferase; TH, b-ketoadipyl-CoA thiolase; ChnD, 6-hydroxycaproate dehydrogenase; ChnE, 6-oxohexanoic dehydrogenase; ACoAL, adipate-CoA ligase; ACoAT, acetyl-CoA C-acyltransferase [54].
3. The Application of Bacteria of the Genus Bacillus to Materials’ Biodamage Control
It should be noted that today the question of the application of biological control for protection against corrosion damage is debatable, as none of the methods for corrosion inhibition by microorganisms found to be successful reached the practical application stage [7]. However, there are noteworthy publications on the possibility of using bacteria of the genus Bacillus to control the biodamage of materials.
The inhibition of corrosion by bacteria is often accomplished by: (1) a decrease in the corrosive action of the medium in restricted parts of the metal–bulk solution interface, for instance, by neutralizing the acidity of the medium; (2) the formation of protective films on the metal surface or providing new protective films, in particular through the production of EPS with metal-binding abilities; and (3) decreasing the cathodic reaction due to the consumption of a cathodic electron acceptor by microbes [6].
In addition, the biocontrol properties of some microorganisms are determined by their high antibacterial properties associated with the formation of antimicrobial compounds [55] and antibiofilm-forming properties due to their ability to affect the quorum sensing system [56]. Such microorganisms include members of the genus Bacillus. Thus, a number of studies have proved that corrosion results from the formation of biofilms, while Bacillus sp. [57][58], B. thuringiensis strain SN8 [59], B. cereus [60], B. licheniformis (including genetically modified ones) [61], B. subtilis [62][63][64], B. firmus strain H2O-1 [65][66] and B. brevis [62] are responsible for producing antimicrobial compounds.
In particular, in conditions of artificial seawater in the presence of a protective biofilm of B. subtilis strain WB600, passivation of Al 2024 was recorded. The addition of antibiotics led to the death of bacteria in the biofilm, which was reflected in the impedance spectra. Adding antibiotics to sterile artificial seawater had no effect on corrosion processes [64].
The inhibiting effect of B. thuringiensis strain SN8 on steel is shown in the publication by Bano et al. [59]. B. thuringiensis strain SN8 inhibited the MIC of materials in soil conditions over a period of 150 days. The corrosion-preventing properties of the B. thuringiensis strain SN8 dissipated after this period. It stands to reason that corrosion-inhibiting/protective properties of biological agents must be verified in term of the maximum time frame for their effectiveness [67].
The inhibitory effect of a biofilm formed by B. cereus was shown by Aїmeur et al. [60].
Ornek et al. evaluated the effect of aluminum-chelating anionic peptides, which release biofilms of natural and genetically modified Bacillus on the pitting of aluminum alloy in continuous reactors [61]. The corrosion rate of aluminum alloy 2024 was reduced by 90% due to γ-polyglutamate-forming biofilm of B. licheniformis. Meanwhile, one of the peptides that secreted the genetically engineered biofilm of B. subtilis only slightly reduced the pitting compared to the biofilm on the control material, which already significantly reduces the corrosion of aluminum compared to the sterile control one.
From thermophilic B. licheniformis on a steel substrate, the bacterial biofilm and the nature of its extracellular polymeric substances have been probed chemically and electrochemically for their influences on metal dissolution within an incubation period [68]. Corrosion inhibition in the presence of varying concentrations (in CFU/ml) of these bacteria in biotic inoculate systems is explained in terms of corrosion resistance and biofilm capacity. Steel’s corrosion rate is reduced significantly in saline culture medium within the range of concentrations of bacteria under study compared with the sterile control. This is attributed to the adhesion of relatively compact and dense “beneficial” biofilm as well as the secretion of corrosion-inhibiting substances from the bacterial biofilm as revealed during surface analysis [68].
Jayaraman et al. [62] found that biofilms of genetically engineered B. subtilis secreting the antibiotics indolicidin, bactenecin and probacterin are able to inhibit the growth of SRB that cause corrosion (D. vulgaris and D. gigas) and significantly reduce corrosion in a continuous culture.
The gramicidin S peptide, which naturally produced the biofilm-forming strain of B. brevis, also inhibited the colonization of SRB and reduced the corrosion of the steel [69]. It should be noted that antibiotics, such as ampicillin, inhibited only the growth of SRB when added prior to the colonization of SRB. At the same time, the associative culture of B. brevis (producing S-gramicidin) with SRB completely blocked the growth of the latter.
Bacillus sp. strain H2O-1, isolated from the connate water of a Brazilian reservoir, produces an antimicrobial substance (denoted as AMS H2O-1) that is active against sulfate-reducing bacteria, which are the major bacterial group responsible for biogenic souring and biocorrosion in petroleum reservoirs. AMS H2O-1 is a mixture of four surfactin-like homologues, and its biocidal activity and surfactant properties suggest that this compound may be a good candidate for sulfate-reducing bacterial control. Thus, it is a potential alternative to the chemical biocides or surface-coating agents currently used to prevent SRB growth in petroleum industries [65].
A new antimicrobial substance is being developed by using biosurfactant produced by indigenous oil reservoir Bacillus sp. to prevent and eradicate biofilm [58]. The biosurfactant is able to inhibit Pseudomonas sp. strain 1 and Pseudomonas sp. strain 2 attachment to carbon steel ST37 surfaces and also able to eradicate preformed biofilm on steel surfaces. This study also showed the reduction of corrosion rate in carbon steel ST37 as a result of material treatment with biosurfactants. The authors suggest that the biosurfactant in this study could be a good candidate for a new anticorrosion agent [58].
The effects of bacteria of the genus Bacillus depend not only on the formation of antimicrobial compounds and protective biofilm, but also on the type of damaged material. In particular, Juzeliunas and co-authors found that B. mycoides accelerates zinc corrosion, inhibits aluminum corrosion and does not affect mild steel [70].
Bacillus sp. strain ПКИ-12Л is considered to be a promising biological agent for the creation of a biological product for the protection of polymeric composite materials from biological damage due to its fungicidal activity [71].
The synthesis of extracellular molecules such as biosurfactants or proteins should have major influence on bacterial adhesion. These molecules may be adsorbed on surfaces and modify their energy characteristics. Under these conditions, adhesion rates are not dependent upon the initial characteristics of the substrate but on those of fouled surfaces [72].
Lipopeptides produced by B. subtilis (surfactin, iturin and fengycin) are able to modify the surface hydrophobicity of stainless steel and Teflon and, consequently, influence microbial adhesion to their surfaces [73]. The authors showed that these effects depend on the lipopeptide type, the concentration and the substratum. Thus, iturin A had no effect in relation to stainless steel; surfactin and fengycin increased the hydrophobicity of steel. The bioconditioning of surfaces using microbial surfactants has been suggested as a new strategy for reducing adhesion of bacteria to surfaces [73].
Among the representatives of the genus Bacillus as controlling agents of biodamage, the bacterium B. velezensis deserves attention [56][55]. It should be noted that the current heterotypic synonyms of B. velezensis are B. amyloliquefaciens subsp. plantarum, B. methylotrophicus, “B. oryzicola”, and B. methylotrophicus subsp. plantarum [74][75].
In the article of Wang et al. [76], high activity against marine fouling by proteases isolated from B. velezensis strain SM-1 was noted. This may be the basis of the modern eco-friendly approach to creating materials against fouling. Khan et al. isolated the bacterium B. methylotrophicus WY with the ability to quench the quorum, which can be used to control membrane biofouling [56]. In particular, it was discovered that this strain has significant influence on the degradation of acylhomoserinlactone (AHSL)—a kind of signaling molecule required for the development of biofilm. The inhibitory activity of B. velezensis strain K68 against the biofilm of Streptococcus mutans on polystyrene was studied [77]. Bacillus sp. strain H2O-1 (with 99.8 and 99.5% similarity of the 16S rRNA gene sequence between it and the type strains of B. amyloliquefaciens and B. methylotrophicus, respectively) produced an antimicrobial substance that was active against sulfate-reducing bacteria [65].
References
- Andreyuk, K.; Kozlova, I.; Kopteva, Z.; Pilyashenko-Novokhatny, A.; Zanina, V.; Purish, L. Mikrobna Koroziia Pidzemnykh Sporud; Naukova Dumka Publishing House: Kyiv, Ukraine, 2005; 258p. (In Ukrainian)
- Tregubova, N.V.; Kochkarov, R.H.; Morgunova, A.V.; Drizhd, N.A.; Dinaev, E.K. Biopovrezhdeniya Produktov; Publishing and Information Center “Fabula”: Stavropol, Russia, 2019; 100p. (In Russian)
- Netrusov, A.I.; Bonch-Osmolovskaya, E.A.; Gorlenko, V.M.; Ivanov, M.V.; Karavajko, G.I.; Kozhevin, P.A.; Kolotilova, N.N.; Kotova, I.B.; Maksimov, V.N.; Nozhevnikova, A.N.; et al. Ekologiya Mikroorganizmov; Netrusov, A.I., Ed.; Academia Publishing Center: Moscow, Russia, 2004; 272p. (In Russian)
- Scarascia, G.; Wang, T.; Hong, P.-Y. Quorum sensing and the use of quorum quenchers as natural biocides to inhibit sulfate-reducing bacteria. Antibiotics 2016, 5, 39.
- Smykun, N.V. Rozvytok koroziino-nebezpechnoi mikroflory gruntu pid vplyvom deiakykh pestytsydiv. Mikrobiol. Z. 2008, 70, 74–87. (In Ukrainian)
- Lin, J.; Ballim, R. Biocorrosion control: Current strategies and promising alternatives. Afr. J. Biotechnol. 2012, 11, 15736–15747.
- Little, B.J.; Blackwood, D.J.; Hinks, J.; Lauro, F.M.; Marsili, E.; Okamoto, A.; Rice, S.A.; Wade, S.A.; Flemming, H.-C. Microbially influenced corrosion—Any progress? Corros. Sci. 2020, 170, 108641.
- Hiraga, K.; Taniguchi, I.; Yoshida, S.; Kimura, Y.; Oda, K. Biodegradation of waste PET: A sustainable solution for dealing with plastic pollution. EMBO Rep. 2019, 20, e49365.
- Restrepo-Flórez, J.-M.; Bassi, A.; Thompson, M.R. Microbial degradation and deterioration of polyethylene—A review. Int. Biodeter. Biodegr. 2014, 88, 83–90.
- Ghatge, S.; Yang, Y.; Jae-Hyung Ahn, J.-H.; Hur, H.-G. Biodegradation of polyethylene: A brief review. Appl. Biol. Chem. 2020, 63, 27.
- Hakkarainen, M.; Albertsson, A. Environmental degradation of polyethylene. Adv. Polym. Sci. 2004, 169, 177.e199.
- Novotný, Č.; Malachova, K.; Adamusc, G.; Kwiecień, M.; Lotti, N.; Soccio, M.; Verney, V.; Fava, F. Deterioration of irradiation/high-temperature pretreated, linear low-density polyethylene (LLDPE) by Bacillus amyloliquefaciens. Int. Biodeterior. Biodegrad. 2018, 132, 259–267.
- Leja, K.; Lewandowicz, G. Polymer Biodegradation and Biodegradable Polymers—A Review. Pol. J. Environ. Stud. 2010, 19, 255–266.
- Van Beilen, J.B.; Li, Z.; Duetz, W.A.; Smits, T.H.M.; Witholt, B. Diversity of Alkane Hydroxylase Systems in the Environment. Oil Gas Sci. Technol.-Rev. IFP 2003, 58, 427–440.
- Seneviratne, G.; Tennkoon, N.S.; Weerasekara, M.L.M.A.W.; Nandasena, K.A. Polyethylene biodegradation by a developed Penicillium-Bacillus biofilm. Curr. Sci. 2006, 90, 20–21.
- Montazer, Z.; Habibi Najafi, M.B.; Levin, D.B. Challenges with Verifying Microbial Degradation of Polyethylene. Polymers 2020, 12, 123.
- Das, M.P.; Kumar, S. An approach to low-density polyethylene biodegradation by Bacillus amyloliquefaciens. 3 Biotech 2015, 5, 81–86.
- Wasserbauer, R.; Beranova, M.; Vancurova, D.; Dolezel, B. Biodegradation of polyethylene foils by bacterial and liver homogenates. Biomaterials 1990, 11, 36–40.
- Sowmya, H.V.; Ramalingappa, M.K.; Thippeswamy, B. Biodegradation of polyethylene by Bacillus cereus. Adv. Polym. Sci. Technol.-Int. J. 2014, 4, 28–32.
- Suresh, B.; Maruthamuthu, S.; Palanisamy, N.; Ragunathan, R.; Pandiyaraj, K.N.; Muralidharan, V.S. Investigation on biodegradability of polyethylene by Bacillus cereus strain Ma-Su isolated from compost soil. Int. Res. J. Microbiol. 2011, 2, 292–302.
- Muhonja, C.N.; Makonde, H.; Magoma, G.; Imbuga, M. Biodegradability of polyethylene by bacteria and fungi from Dandora dumpsite Nairobi-Kenya. PLoS ONE 2018, 13, e0198446.
- Abdullahi, M.; Saidu, B. Biodegradation of polythene and plastic using fadama soil amended with organic and inorganic fertilizer. Indian J. Sci. Res. 2013, 4, 17–24.
- Vimala, P.P.; Mathew, L. Biodegradation of polyethylene using Bacillus subtilis. Proc. Technol. 2016, 24, 232–239.
- Ibiene, A.A.; Stanley, H.O.; Immanuel, O.M. Biodegradation of Polyethylene by Bacillus sp. Indigenous to the Niger Delta Mangrove Swamp. Nig. J. Biotech. 2013, 26, 68–79.
- Waqas, M.; Haris, M.; Asim, N.; Islam, H.; Abdullah, A.; Khan, A.; Khattak, H.; Waqas, M.; Ali, S. Biodegradable potential of Bacillus amyloliquefaciens and Bacillus safensis using low density polyethylene thermoplastic (LDPE) substrate. Eur. J. Environ. Public Health 2021, 5, em0069.
- Pinchuk, L.S.; Makarevich, A.V.; Vlasova, G.M.; Kravtsov, G.A.; Shapovalov, V.A. Electret-thermal analysis to assess biodegradation of polymer composites. Int. Biodeter. Biodegr. 2004, 54, 13–18.
- Sudhakar, M.; Doble, M.; Sriyutha Murthy, P.; Venkatesan, R. Marine microbe-mediated biodegradation of low- and high-density polyethylenes. Int. Biodeterior. Biodegrad. 2008, 61, 203–213.
- Abrusci, C.; Pablos, J.L.; Marín, I.; Espí, E.; Corrales, T.; Catalina, F. Comparative effect of metal stearates as pro-oxidant additives on bacterial biodegradation of thermal- and photo-degraded low density polyethylene mulching films. Int. Biodeterior. Biodegrad. 2013, 83, 25–32.
- Yang, J.; Yang, Y.; Wu, W.-M.; Zhao, J.; Jiang, L. Evidence of Polyethylene Biodegradation by Bacterial Strains from the Guts of Plastic-Eating Waxworms. Environ. Sci. Technol. 2014, 48, 13776–13784.
- Usha, R.; Sangeetha, T.; Palaniswamy, M. Screening of polyethylene degrading microorganisms from garbage soil. Libyan Agric. Res. Cent. J. Int. 2011, 2, 200–204.
- Kumari, A.; Chaudhary, D.R.; Jha, B. Destabilization of polyethylene and polyvinylchloride structure by marine bacterial strain. Environ. Sci. Pollut. Res. 2019, 26, 1507–1516.
- Aravinthan, A.; Arkatkar, A.; Juwarkar, A.A.; Doble, M. Synergistic growth of Bacillus and Pseudomonas and its degradation potential on pretreated polypropylene. Prep. Biochem. Biotechnol. 2016, 46, 109–115.
- Auta, H.S.; Emenike, C.U.; Fauziah, S.H. Screening for Polypropylene Degradation Potential of Bacteria Isolated from Mangrove Ecosystems in Peninsular Malaysia. Int. J. Biosci. Biochem. Bioinform. 2017, 7, 245–251.
- Kimi, J.; Bhunia, H.; Reddy, M.S. Degradation of polypropylene-poly-L-lactide blends by Bacillus isolates: A microcosm and field evaluation. Bioremed. J. 2021, 1–15.
- Auta, H.S.; Emenike, C.U.; Fauziah, S.H. Screening of Bacillus strains isolated from mangrove ecosystems in Peninsular Malaysia for microplastic degradation. Environ. Pollut. 2017, 231, 1552–1559.
- Mohanan, N.; Montazer, Z.; Sharma, P.K.; Levin, D.B. Microbial and Enzymatic Degradation of Synthetic Plastics. Front. Microbiol. 2020, 11, 2837.
- Shimpi, N.; Mishra, S.; Kadam, M. Biodegradation of polystyrene (PS)-poly(lactic acid) (PLA) nanocomposites using Pseudomonas aeruginosa. Macromol. Res. 2012, 20, 181–187.
- Atiq, N.; Ahmed, S.; Ishtiaq Ali, M.; Andleeb, S.; Ahmad, B.; Robson, G. Isolation and identification of polystyrene biodegrading bacteria from soil. Afr. J. Microbiol. Res. 2010, 4, 1537–1541.
- Mohan, A.J.; Sekhar, V.C.; Bhaskar, T.; Nampoothiri, K.M. Microbial assisted High Impact Polystyrene (HIPS) degradation. Bioresour. Technol. 2016, 213, 204–207.
- Meng, T.K.; Beng, D.Y.Y.; Mohd Kassim, A.S.; Razak, A.H.A.; Mohd Fauzi, N.A. Optimization of Polystyrene Biodegradation using Response Surface Methodology (RSM) Measured by Simple Colorimetric Method. Int. J. Eng. Technol. 2018, 7, 216–220.
- Asmita, A.; Shubhamsingh, T.; Tejashree, S. Isolation of plastic degrading micro-organisms from soil samples collected at various locations in Mumbai, India. Int. Res. J. Environ. Sci. 2015, 4, 77–85.
- Ganesh Kumar, A.; Hinduja, M.; Sujitha, K.; Nivedha Rajan, N.; Dharani, G. Biodegradation of polystyrene by deep-sea Bacillus paralicheniformis G1 and genome analysis. Sci. Total Environ. 2021, 774, 145002.
- Lou, Y.; Ekaterina, P.; Yang, S.-S.; Lu, B.; Liu, B.; Ren, N.; Corvini, P.F.X.; Xing, D. Biodegradation of polyethylene and polystyrene by greater wax moth larvae (Galleria mellonella L.) and the effect of co-diet supplementation on the core gut microbiome. Environ. Sci. Technol. 2020, 54, 2821–2831.
- Dąbrowska, G.B.; Janczak, K.; Richert, A. Combined use of Bacillus strains and Miscanthus for accelerating biodegradation of poly(lactic acid) and poly(ethylene terephthalate). PeerJ 2021, 9, e10957.
- Vague, M.; Chan, G.; Roberts, C.; Swartz, N.A.; Mellies, J. Pseudomonas isolates degrade and form biofilms on polyethylene terephthalate (PET) plastic. BioRxiv 2019, 647321.
- Wei, R.; Zimmermann, W. Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: How far are we? Microb. Biotechnol. 2017, 10, 1308–1322.
- Giacomucci, L.; Raddadi, N.; Soccio, M.; Lotti, N.; Fava, F. Polyvinyl chloride biodegradation by Pseudomonas citronellolis and Bacillus flexus. New Biotechnol. 2019, 52, 35–41.
- Howard, G.T. Polyurethane Biodegradation. In Microbial Degradation of Xenobiotics, Environmental Science and Engineering; Singh, S.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 371–394.
- Tan, J.D.; Ohwada, T. Isolation and identification of microorganisms for polyurethane degradation. Ann. Trop. Res. 2019, 41, 57–66.
- Rowe, L.; Howard, G.T. Growth of Bacillus subtilis on polyurethane and the purification and characterization of a polyurethanase-lipase enzyme. Int. Biodeter. Biodegr. 2002, 50, 33–40.
- Shah, Z.; Krumholz, L.; Aktas, D.F.; Hasan, F.; Khattak, M.; Shah, A.A. Degradation of polyester polyurethane by a newly isolated soil bacterium, Bacillus subtilis strain MZA-75. Biodegradation 2013, 24, 865–877.
- Nakkabi, A.; Sadiki, M.; Fahim, M.; Ittobane, N.; Ibnsouda Koraichi, S.; Barkai, H.; El abed, S. Biodegradation of Poly(ester urethane)s by Bacillus subtilis. Int. J. Environ. Res. 2015, 9, 157–162.
- Mahajan, N.; Gupta, P. New Insights into the Microbial Degradation of Polyurethanes. RSC Adv. 2015, 5, 41839–41854.
- Ru, J.; Huo, Y.; Yang, Y. Microbial Degradation and Valorization of Plastic Wastes. Front. Microbiol. Rev. 2020, 11, 442.
- Chowdhury, S.P.; Hartmann, A.; Gao, X.W.; Borriss, R. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42—A review. Front. Microbiol. 2015, 6, 780.
- Khan, R.; Shen, F.; Khan, K.; Liu, L.X.; Wu, H.H.; Luo, J.Q.; Wan, Y.H. Biofouling control in membrane filtration system by newly isolated novel quorum quenching bacterium, Bacillus methylotrophicus sp. WY. RSC Adv. 2016, 6, 28895–28903.
- Du, J.; Li, S.; Liu, J.; Yu, M. Corrosion behavior of steel Q235 co-influenced by Thiobacillus thiooxidans and Bacillus. J. Beijing Univ. Aeronaut. Astronaut. 2014, 40, 31–38.
- Purwasena, I.A.; Astuti, D.I.; Fauziyyah, N.A.; Putri, D.A.S.; Sugai, Y. Inhibition of microbial influenced corrosion on carbon steel ST37 using biosurfactant produced by Bacillus sp. Mater. Res. Express. 2019, 6, 115405.
- Bano, A.S.; Qazi, J.I. Soil buried mild steel corrosion by Bacillus cereus—SNB4 and its inhibition by Bacillus thuringiensis—SN8. Pak. J. Zool. 2011, 43, 555–562.
- Aїmeur, N.; Houali, K.; Hamadou, L.; Benbrahim, N.; Kadri, A. Influence of strain Bacillus cereus bacterium on corrosion behaviour of carbon steel in natural sea water. Corros. Eng. Sci. Technol. 2015, 50, 579–588.
- Ornek, D.; Jayaraman, A.; Syrett, B.C.; Hsu, C.H.; Mansfeld, F.B.; Wood, T.K. Pitting corrosion inhibition of aluminum 2024 by Bacillus biofilms secreting polyaspartate or g-polyglutamate. Appl. Microbiol. Biotechnol. 2002, 58, 651–657.
- Jayaraman, A.; Mansfeld, F.B.; Wood, T.K. Inhibiting sulfate-reducing bacteria in biofilms by expressing the antimicrobial peptides indolicidin and bactenecin. J. Ind. Microbiol. Biotechnol. 1999, 22, 167–175.
- Wadood, H.Z.; Rajasekar, A.; ·Ting, Y.-P.; Sabari, A.N. Role of Bacillus subtilis and Pseudomonas aeruginosa on Corrosion Behaviour of Stainless Steel. Arab. J. Sci. Eng. 2015, 40, 1825–1836.
- Zuo, R.; Kus, E.; Mansfeld, F.; Wood, T.K. The importance of live biofilms in corrosion protection. Corros. Sci. 2005, 47, 279–287.
- Korenblum, E.; de Araujo, L.V.; Guimarães, C.R.; de Souza, L.M.; Sassaki, G.; Abreu, F.; Nitschke, M.; Lins, U.; Freire, D.M.G.; Barreto-Bergter, E.; et al. Purification and characterization of a surfactin-like molecule produced by Bacillus sp. H2O-1 and its antagonistic effect against sulfate reducing bacteria. BMC Microbiol. 2012, 12, 252.
- Korenblum, E.; Sebastián, G.V.; Paiva, M.M.; Coutinho, C.M.L.M.; Magalhães, F.C.M.; Peyton, B.M.; Seldin, L. Action of antimicrobial substances produced by different oil reservoir Bacillus strains against biofilm formation. Appl. Microbiol. Biotechnol. 2008, 79, 97–103.
- Hussain, A.; Bano, S.S.; Qazi, J.I. Corrosion of Mild Steel Simulating Long Term Soil Burial Field Conditions Differing in Nutritional and Biotic Components. World Appl. Sci. J. 2013, 22, 985–990.
- Eduok, U.; Khaled, M.; Khalil, A.; Suleiman, R.; El Ali, B. Probing the corrosion inhibiting role of thermophilic Bacillus licheniformis biofilm 1 on steel in a saline axenic culture. RSC Adv. 2016, 6, 18246–18256.
- Jayaraman, A.; Hallock, P.J.; Carson, R.M.; Lee, C.C.; Mansfeld, F.B.; Wood, T.K. Inhibiting sulfate-reducing bacteria in biofilms on steel with antimicrobial peptides generated in situ. Appl. Microbiol. Biot. 1999, 52, 267–275.
- Juzeliunas, E.; Ramanauskas, R.; Lugauskas, A.; Leinartas, K.; Samulevicene, M.; Sudavicius, A. Influence of wild strain Bacillus mycoides on metals: From corrosion acceleration to environmentally friendly protection. Electrochim. Acta 2006, 51, 6085–6090.
- Erofeevskaya, L.A.; Kychkin, A.K.; Kychkin, A.A. Prognozirovanie biodegradacii polimernyh kompozicionnyh materialov v klimaticheskih usloviyah Yakutii. Innovacii Invest. 2019, 6, 202–207. (In Russian)
- Garry, P.; Vendeuvre, J.L.; Bellon-Fontaine, M.N. Surface properties and adhesion of Bacillus cereus and Bacillus subtilis to polyurethane—Influence of growth temperature. J. Disper. Sci. Technol. 1998, 19, 1175–1197.
- Shakerifard, P.; Gancel, F.; Jacques, P.; Faille, C. Effect of different Bacillus subtilis lipopeptides on surface hydrophobicity and adhesion of Bacillus cereus 98/4 spores to stainless steel and Teflon. Biofouling 2009, 25, 533–541.
- NCBI. Taxonomy Browser. Available online: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=492670&lvl=3&lin=f&keep=1&srchmode=1&unlock (accessed on 5 September 2019).
- Dunlap, C.A.; Kim, S.-J.; Kwon, S.-W.; Rooney, A.P. Bacillusvelezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and ‘Bacillus oryzicola’ are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. Int. J. Syst. Evol. Microbiol. 2016, 66, 1212–1217.
- Wang, L.; Yu, L.; Lin, C. Extraction of protease produced by sea mud bacteria and evaluation of antifouling performance. J. Ocean Univ. China 2019, 18, 1139–1146.
- Yoo, Y.; Seo, D.-H.; Lee, H.; Cho, E.-S.; Song, N.-E.; Nam, T.G.; Nam, Y.-D.; Seo, M.-J. Inhibitory effect of Bacillus velezensis on biofilm formation by Streptococcus mutans. J. Biotechnol. 2019, 298, 57–63.
- Karn, S.K.; Fang, G.; Duan, J. Bacillus sp. acting as dual role for corrosion induction and corrosion inhibition with carbon steel (CS). Front. Microbiol. 2017, 24, 2038.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.9K
Revisions:
3 times
(View History)
Update Date:
12 Nov 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




