Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Abid Khan | + 2490 word(s) | 2490 | 2021-10-21 04:51:46 | | | |
| 2 | Catherine Yang | Meta information modification | 2490 | 2021-10-29 07:40:18 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Khan, A. Plant Heat Shock Proteins. Encyclopedia. Available online: https://encyclopedia.pub/entry/15534 (accessed on 07 February 2026).
Khan A. Plant Heat Shock Proteins. Encyclopedia. Available at: https://encyclopedia.pub/entry/15534. Accessed February 07, 2026.
Khan, Abid. "Plant Heat Shock Proteins" Encyclopedia, https://encyclopedia.pub/entry/15534 (accessed February 07, 2026).
Khan, A. (2021, October 29). Plant Heat Shock Proteins. In Encyclopedia. https://encyclopedia.pub/entry/15534
Khan, Abid. "Plant Heat Shock Proteins." Encyclopedia. Web. 29 October, 2021.
Copy Citation
Plant heat shock proteins (HSPs), as chaperones, play a pivotal role in conferring biotic and abiotic stress tolerance. Moreover, HSP also enhances membrane stability and detoxifies the reactive oxygen species (ROS) by positively regulating the antioxidant enzymes system.
heat shock factor
biotic stress
abiotic stress
protein folding
stress resistance
1. Introduction
Plants are sessile organisms and are subjected to various threats, both biotic and abiotic. These stresses, individually or in combination, result in huge losses in terms of growth, development, and yield and sometimes threaten the survival of the plant [1]. Plants continuously confront harsh environments like high/low temperatures, drought, salts, heavy metals, light, flooding and physical wounding [2][3][4][5]. Biotic stresses like pathogens (Viruses, Bacteria, Fungi) and pests such as nematodes, insects, and rodents also restrict plant productivity [6][7][8][9][10]. Negative effects of these stresses on the plant germination [11], are stunted growth [12][13], sunburn and scorching of leaves [14], loss of photosynthetic pigment, decreased production of photo-assimilates, and depletion of carbohydrate reserves which results in starvation [15][16][17][18]. Abiotic stresses also negatively affect the reproductive characteristics of plants by enhancing male sterility [19] and increasing premature flower and fruit drop [20] which results in significant low yield and quality. It has been reported that the increase in temperature by 1 °C results in a 4–10% yield decrease [21]. As a consequence of these stresses, reactive oxygen species (ROS) are produced which lead to oxidative stress and, ultimately, results in cell death. ROS could be singlet oxygen (1O2), superoxide radical (O2•−), hydrogen peroxide (H2O2) and hydroxyl radical (OH −), which are produced in cell organelles such as mitochondria, peroxisomes and chloroplasts in oxidative stress situations and react with all types of macromolecules like pigments, proteins, lipids and DNA [22][23].
Plants respond morphologically to elevated temperature and light stress by changing the leaf orientation [13], anatomically by altering stomatal conductance and increased leaf pubescence [24][25], and phenologically by shifting and improvising the developmental stages to escape the abiotic stress condition [26]. Plants also change the metabolic processes and physiology to retain root hydraulic conductance [27], accumulation of the compatible osmolytes, such as sugars, sugar alcohols, proline and phenolic compounds under saline and water-logged conditions, as well as high temperature and water deficit conditions [28]. Moreover, plants manage to maintain photosynthetic machinery [29] by changing their assimilate partitioning a shift occurs from symplastic to apo-plastic [30]. During the onset of the stress situations, plants also improvise the hormonal balance of abscisic acid (ABA), ethylene, and salicylic acid (SA) as a signaling molecule in the systemic acquired resistance. Similarly, jasmonic acid (JA) and other steroids enhance stress tolerance and resistance [31]. Furthermore, secondary metabolites, such as isopropanioid, carotenoid, flavonoid, anthocyanin, lignin, and isoprenoids [30][32], also are produced and accumulated.
Besides these adaptations, plants also have sophisticated adaptive systems at the cellular and molecular levels. During the onset of stress, plants reduce the synthesis of normal protein production, and transcribe and translate heat shock proteins (HSPs). Added to transcriptional regulations, plants also have some sophisticated post-transcriptional modifications which help the plant to cope with these stresses, such as alternative splicing and micro RNA (miRNA). Alternative splicing, which generates multiple copies from a single gene, helps the plants to mitigate abiotic stresses [33]. One of the important plant post-transcriptional modification strategies is the miRNA, which binds to the mRNA at any point to repress translation or direct cleavage of the mRNA. Some of the miRNAs also are involved with abiotic stress tolerance [34].
2. Occurrence of HSP in Plants
The heat shock response is not unique in plants. It was first discovered in the early 1960s by an Italian scientist, F. Ritossa, in Drosophila melanogaster, while working on high-temperature stress [35][36]. HSPs were studied in Saccharomyces cerevisiae, by McAlister et al. (1980) [37], Escherichia coli, by Yamamori et al. (1982) [38], and plants (Glycine max), Lin et al. (1984) [39]. A comparison of the response in different organisms has shown that HSPs are conserved highly across organisms [40]. The evolutionary conservation of the heat shock response shows that the production of HSPs is a fundamental and essential process in all organisms [41].
Plants, due to their sessile nature, have evolved different efficient strategies to ensure their generations in the challenging competitive environment. The numbers of stress related genes, including HSPs, are greater in plants than other organisms due to whole genome duplication and gene retention in their evolutionary past [42]. Additionally, plants contain chloroplasts which have their own genome and de novo protein synthesis machinery [43]. A strategy of the plant to cope with stressful situations (Figure 1), is the synthesis of normal protein reduced while the synthesis of stress-related proteins i.e., HSPs, is enhanced.
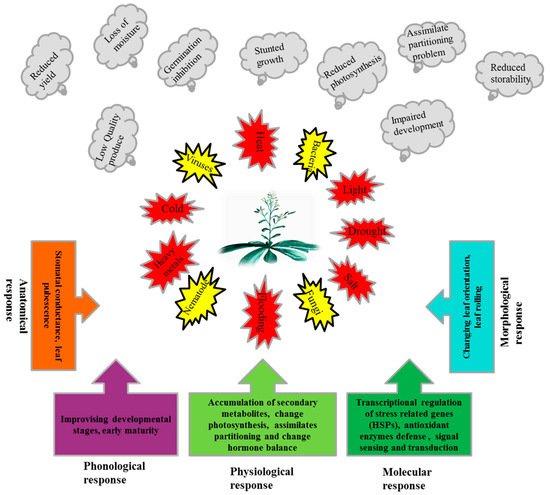
Figure 1. Exposures of plants to different biotic and abiotic stresses, adverse effects of these stresses on plants and response mechanisms of plants to these stresses.
Many HSPs have been reported in a wide range of organisms from prokaryotes to eukaryotes [40][44]. These HSPs across the organisms are conserved highly, with little difference except for HSP33, which differs in plants from that in bacteria [45]. The genes which encode different HSPs are found in different cell compartments, such as the nucleus, mitochondria, chloroplast, endoplasmic reticulum and cytosol [46]. Similarly, the accumulation of these HSPs in different parts of the cell also depends on the intensity of the stress. Nuclear HSPs, for instance, are accumulating in the cytosol at the lower and higher temperatures of 27 °C and 43 °C, respectively, while the same aggregate in chloroplast is at 37 °C [47]. Different HSPs are found and differentially expressed in different species, even in different genotypes but in the same species, as investigated by Korotaeva et al., (2001) and Nieto-Sotelo et al., (2002) [48][49] in small HSPs where five sHSPs showed response to a higher temperature (42 °C) in maize but only one expressed in wheat and rice. Likewise, HSP68 expressed in mitochondria under stress situations in potatoes, tomatoes, and soybeans [50]. Some HSPs showed a tissue-specific response to stress situations; HSP101 was expressed more in reproductive parts like tassels, ear, and endosperm, than in vegetative parts like leaves and roots in maize [51]. Some HSPs responded differently to the varying length of stresses. As Heckathorn et al., (1989) [52] reported, in HSP45, a nuclear protein accumulated in the chloroplast at a 3 h exposure to heat stress, which returned to its native state after removal of stress. Similarly, HSPs triggered differently with different development stages. HSP45, for example, showed a response in the whole plant to a stress situation, while HSP64 and -72 only showed expression in the reproductive parts i.e., pollens [53][54]. It can be deduced that these are the key regulators which show different responses to varying levels of stress in different parts of the plants.
3. Role of HSPs in Plant Defense
Plants are subjected to various biotic and abiotic stresses, singly or in combination, which adversely affect plant growth, development, and survival [55]. To cope with these stresses, plants have evolved various defense strategies i.e., physical [13], anatomical [25], and physiological [27][56]. Plants also respond to stress situations at the molecular level by altering gene expression, synthesis of stress-related biomolecules and proteins including heat stress proteins, to enable plants to ensure their generation in challenging situations.
3.1. Biotic Stress Tolerance
Plant growth, development, yield, and quality are affected adversely by several biotic factors such as pathogenic bacteria, fungi, viruses, and nematodes. Biotic factors, directly deprive their host plants of their nutrients, which result in reduced plant vigor, growth, productivity and sometimes leads to death of the host plants. Biotic stresses are major cause of pre- and post-harvest losses. Animals have an immune system, which helps them to adapt to biotic stresses such as new diseases and memorized the past infections, while plants lack such a system. Although plants lack this adaptive immune system, they have evolved several sophisticated strategies to counteract these biotic stresses [57]. These defense mechanisms are stored in the plant’s genome at the genetic level, which encode thousands of stress resistance genes. One of the adaptive systems plants employ in response to biotic stress is through regulation of HSPs (Table 1). HSP response to biotic stresses depend on the nature of the causal organisms and plant genotypes, either susceptible or resistant, and the developmental stage [6].
Table 1. Summary of the role of plant HSP under biotic stresses.
| Biotic Factor |
Plant | Pathogen | Disease Caused | HSP Response | Expression Pattern |
Reference |
|---|---|---|---|---|---|---|
| Viruses | Arabidopsis thaliana | TVCTV, ORTV, PVX, CMV, and TuMV |
Penstemon disease, red spots on leaves, deformed leaves and stunted growth | HSP17.6 HSP17.4 |
Up/down | [58] |
| Solanum lycopersicum | TYLCV | Stunted and bushy growth and excessive branches, abnormal leaf shapes, curled inward or upward, and flower and fruit drops | HSP70 HSP90 |
Up/down | [59][58][60] | |
| Nicotiana benthamina | CNV, RCNMV, and TMV | Mosaic and discoloration on leaves of a wide-range of the host plant | HSP70 HSP90 |
Up | [61][62][63] | |
| Solanum tuberosum | SYNV and INSV | Yellow and black rings, spots and lesions on leaves, and leads to plant death | HSP18 HSP20 |
Up | [64] | |
| Oryza sativa | RSV and RBSDV | Dark green rigid leaves, white- and sometimes black-streaked strips along the leaves, veins and stem | HSP20 HSP70 |
Up | [7][65][66] | |
| Solanum tuberosum | PVY | Potato tuber necrotic rings, spots, disease and decay | HSP | Up | [67] | |
| Bacteria | Nicotiana benthamina | Ralstonia solanacearum | Wide- range of the host, it enters the xylem of a plant and causes wilting | HSP17 | Up | [68] |
| Ralstonia solani | Bacterial wilt by blockade of conducting vessels | HSP90 | Up | [69] | ||
| Citrus spp | Xanthomonas axonopodis pv. citri | This bacterium causes the citrus canker and spots on leaves and blemishes on fruits. | Hsp15.5 | Up | [70] | |
| Capsicum annuum | Xanthomonas campestris pv. Vesicatoria | Causes leaf and fruit spots on peppers and tomatoes. | Hsp16 HSP20 |
Up | ||
| Arabidopsis thaliana | Pseudomonas syringae | Round to irregular brown spots. These spots enlarge and blight the whole leaves. | HSP17 HSP21 HSP23 |
Down | [71][72] | |
| Fungi | Oryza sativa | Magnaporthe grisea | Causes destructive disease of rice, rice blast, rice seedling blight and pitting disease. | HSP16 HSP17.4 HSP18 |
Up | [73] |
| Solanum lycopersicum | Fusarium oxysporum | Wilting, characterized by clearing of veins, marginal necrosis, yellowing of lower leaves, adventitious roots and ultimate death of tomato plants. | HSP20 | Up | [74] | |
| Rhizopus nigricans Ehrenb. | Rhizopus soft rot first appears as water-soaked areas, which then become sunken and gray mold and dusky black spores grow on the fruit surface of tomatoes. | HSP17.6 | Up | [75] | ||
| Oidium neolycopersici | Fungus that causes white powdery lesions and powdery mildew on tomatoes | HSP72 HSP75 |
Up | [76] | ||
| Arabidopsis thaliana | Rhizoctonia solani | Fungus which causes collar rot, root rot and damping off | HSP17.4 HSP17.6 |
Up | [55][56] | |
| Malus domestica | Venturia inaequalis | Fungus causes scab disease on apples and pears | HSP21 | Down | [77] | |
| Solanum tuberosum | Phytophthora infestans | Fungus causes late blight of potatoes | HSP17.8 HSP70 |
Up Up |
[78] [79] |
|
| Hordeum vulgare | Blumeria graminis f. sp. hordei | Fungus causes powdery mildew on grasses and cereals like barley. | Hsp16.9 Hsp17.5. |
Up | [80] | |
| Nematodes | Gossypium hirsutum | Roylenchulus reniformis | Stunted growth, root necrosis and the plant shows symptoms similar to nutrient and water deficiency | 54 HSP | Up | [7] |
| Glycine max | Meloidogyne javanica | Root knot on tropical crops, it is causes irregular galls and swollen roots | HSP22.4 HSP17.9 HSP17.9 HSP22.4 |
Up | [81][82] | |
| Heterodera glycines | Forms cysts on the roots of soybeans. It causes chlorosis of leaves and stem and root necrosis | HSP20 | Up | [83] | ||
| Helianthus annuus | Meloidogyne incognita | Irregular galls on the root of sunflowers | HSP17.6 HSP17.7 HSP18.6 |
Up | [84][85] | |
| Solanum lycopersicum | Meloidogyne spp. | Knots and galls on the roots of tomatoes, swollen roots and dwarf stem | HSP90 | Up | [86] | |
| Nicotiana benthamina | Meloidogyne incognita | Root knot and galls on tobacco roots and causes wilting of leaves | HSP90 | Up | [87] |
Turnip vein clearing virus (TVCTV), Oilseed rape virus (ORTV), Potato virus X (PVX), Cucumber mosaic virus (CMV), Turnip mosaic virus (TuMV), Tomato yellow leaf curl virus (TYLCV), Cucumber Necrosis Virus (CNV), Red clover necrotic mosaic virus (RCNMV), Tobacco mosaic virus (TMV), Sonchus yellow net virus (SYNV), Impatiens necrotic spot virus (INSV), Rice stripe virus (RSV), Rice black Streaked dwarf Virus (RBSDV), Potato virus Y (PVY).
3.2. Abiotic Stress Tolerance
Abiotic stresses are extreme environmental conditions like extreme temperatures, water deficit, and ion imbalance due to heavy metals and salinity, which pose a serious threat to plants survival, yield and quality. Global warming and climate change, due to industrialization, has further worsened the situation. Greenhouse gases, particularly the concentration of CO2, is increasing constantly in the atmosphere, which is estimated to reach 520 ppm from 410 by the year 2100 [88]. Twenty per cent of the world cultivated land and almost 50% of irrigated land is affected by salinity, and yield loss up to 50% due to drought is projected by the year 2050 [89]. The world population by the year 2050 is approximated to be 9 billion [90], so, in such a situation, biotic and abiotic stress-tolerant cultivars need to be developed using transgenic and omic techniques. The role of HSPs has been studied by various scientists under different abiotic stresses (Table 2).
Table 2. Summary of studies of plant HSP and abiotic stresses.
| Abiotic Factors | Plant | Type of HSP | Expression Pattern | Technique Used | Reference |
|---|---|---|---|---|---|
| High temperature stress | Wheat | HSP70 | up | qRT-PCR | [91] |
| HSP26 | up | qRT-PCR | [92] | ||
| Rice | HSP100 | up | WB | [93] | |
| HSP90 | up | q-PCR | [94][95][96] | ||
| Maize | HSP101 | up | SDS-PAGE | [40][43] | |
| HSP70, HSP17.6 | up | SDS-PAGE | [97] | ||
| Arabidopsis | HSP101 | up | qRT-PCR | [98][89][90][99] | |
| HSP100 | up | SDS-PAGE | [43] | ||
| HSP90 | up | qRT-PCR, WB | [100][101] | ||
| HSP70 | up | qRT-PCR | [102] | ||
| HSP60 HSP70 |
up | qRT-PCR | [103] | ||
| Potato | HSP70 | up | qRT-PCR | [104] | |
| Tomato | HSP70 | up | SDS-PAGE, WB | [105] | |
| HSP20 | up/down | qRT-PCR | [106] | ||
| Pea | HSP17.9 HSP18.1 |
up | qRT-PCR | [107] | |
| HSP70 | up | qRT-PCR | [108] | ||
| Pepper | HSP70 | up/down | qRT-PCR | [109] | |
| HSP70 | up | qRT-PCR | [110] | ||
| HSP60 | up | qRT-PCR | [111] | ||
| HSP20 | up/down | qRT-PCR | [112] | ||
| HSP16.4 | up | qRT-PCR | [113] | ||
| Soybean | HSP90 | up | qRT-PCR | [95] | |
| Cabbage | HSP70 | up | qRT-PCR | [114] | |
| Tea | All HSPs | up | qRT-PCR | [115] | |
| Witch grass | HSP70 | up | MA, qRT-PCR | [116] | |
| Alfalfa | HSP70 | up | qRT-PCR | [117] | |
| Foxtail millet | HSP20 | down | qRT-PCR | [118] | |
| Chrysanthemum | HSP70 | up | MS, qRT-PCR | [119] | |
| Low temperature stress | Arabidopsis | HSP70 | up | MS, qRT-PCR | [120] |
| Tobacco | HSP70 | up | MS, qRT-PCR | [121][122] | |
| Maize | HSP70 | up | MA, qRT-PCR | [123] | |
| Wheat | HSP70 | up | MS | [124] | |
| HSP90 | up | MS | [125] | ||
| HSP60 HSP21 |
down | MS, qRT-PCR | [126] | ||
| Rice | HSP75 HSP95 |
up | MS, qRT-PCR | [127] | |
| HSP90 | up | MS | [128] | ||
| Barley | HSP70 | up | MS | [126] | |
| Chicory | All HSPs | up | MS | [129] | |
| Rape seed | HSP90 | up | qRT-PCR | [130] | |
| Poplar | HSP70 HSP90 |
up | MS | [131] | |
| Pea | HSP70 | up | MS | [132] | |
| Sunflower | HSP60 HSP21 |
down | MS, qRT-PCR | [133] | |
| Tomato | HSP110 HSP70 |
up | qRT-PCR | [134] | |
| Plum | HSP20 | up | qRT-PCR | [135] | |
| Grape | HSP18 HSP22 |
up | qRT-PCR | [136] | |
| Salinity stress | Wheat | HSP70 | up | MS | [137] |
| All HSPs | up | [138] | |||
| Rice | HSP70 | up | qRT-PCR | [139][140] | |
| HSP40 | up | qRT-PCR | [122] | ||
| ClpD1 | up | qRT-PCR | [141] | ||
| Arabidopsis | HSP90.2 HSP90.5 HSP90.7 |
up | qRT-PCR | [142] [95] |
|
| Rose | 17.8 | up | qRT-PCR | [16] | |
| Soybean | HSP90 HSP70 HSP60 HSP20 |
up/down | MS | [143] [144] |
|
| Poplar | HSP100 HSP90 HSP70 HSP60 HSP40 HSP20 |
up | qRT-PCR | [145] | |
| Drought stress | Arabidopsis | HSP70 | up | qRT-PCR | [146] |
| Tobacco | HSP70 BiPs |
up | qRT-PCR | [147] [148] |
|
| Barley | HSP17.5 | up | qRT-PCR | [149] | |
| Rice | HSP70 | up | MA, MS, qRT-PCR | [150] | |
| HSP101 | up | MS | [151] | ||
| HSP17.7 | up | qRT-PCR | [152] | ||
| Maize | HSP70 HSP26 |
up | MS | [153] | |
| Pepper | HSP16.4 | up | qRT-PCR | [113] | |
| Chickpea | HSP70 | up | MS, qRT-PCR | [154] | |
| Sugarcane | HSP70 | up | qRT-PCR | [155] | |
| Cotton | All HSPs | up | qRT-PCR, WB | [156] | |
| Kentucky grass | HSP70 | up | MS | [157] | |
| Poplar | HSP70 | up | MS | [158] | |
| Light stress | Arabidopsis | HSP70 | up | qRT-PCR | [159][160] |
| Goose foot plant | HSP23 | up | SDS-PAGE | [48] | |
| Chlamydomonas | HSP70 | up | MA | [161] | |
| Heavy metal stress | Tomato | HSP70 | up | MS, SDS-PAGE | [162] |
| Rice | HSP70 BiPs |
up | MS, SDS-PAGE | [163] | |
| HSP80 HSP17.9 |
up | MA | [164] | ||
| Poplar | HSP70 | up | qRT-PCR | [156] | |
| Soybean | HSP26 | up | qRT-PCR | [165] | |
| Carrot | HSP17.7 | up | qRT-PCR | [166] | |
| Flaxseed | HSP70 | up | MA, MS | [167] | |
| HSP80 | down | ||||
| Arabidopsis | HSP70 | up | qRT-PCR, NB, MS | [168] | |
| Bird foot trefoil | HSP90 | up | qRT-PCR | [169] | |
| Flooding stress | Arabidopsis | HSP101 HSP70 |
up | qRT-PCR | [170] |
| Tomato | HSP23.6 | up | qRT-PCR | [171] | |
| Rice | HSP70 | up | qRT-PCR | [172] | |
| Maize | HSP70 | up | qRT-PCR, WB | [173] | |
| Soybean | HSP70 | up | qRT-PCR, SDS-PAGE | [174] | |
| HSP60 | up | MS | [175] |
Quantitative real-time polymerase chain reaction (qRT-PCR), Mass spectrometry (MS), Microarray (MA), Western blotting (WB), Northern blotting (NB), Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE).
References
- Mittler, R. Abiotic Stress, the Field Environment and Stress Combination. Trends Plant Sci. 2006, 11, 15–19.
- Swindell, W.R.; Huebner, M.; Weber, A.P. Transcriptional Profiling of Arabidopsis Heat Shock Proteins and Transcription Factors Reveals Extensive Overlap Between Heat and Non-Heat Stress Response Pathways. BMC Genom. 2007, 8, 125.
- Al-Whaibi, M.H. Plant Heat-Shock Proteins: A Mini Review. J. King Saud Univ. Sci. 2011, 23, 139–150.
- Guo, M.; Liu, J.; Ma, X.; Luo, D.; Gong, Z.; Lu, M. The Plant Heat Stress Transcription Factors (HSFs): Structure, Regulation, and Function in Response to Abiotic Stresses. Front. Plant Sci. 2016, 7, 114.
- Xu, Y.; Zhan, C.; Huang, B. Heat Shock Proteins in Association with Heat Tolerance in Grasses. Int. J. Proteom. 2011, 2011, 529648.
- Dodds, P.; Rathjen, J. Plant Immunity: Towards an Integrated View of Plant-Pathogen Interactions. Nat. Rev. Genet. 2010, 11, 539.
- Li, R.; Rashotte, A.M.; Singh, N.K.; Lawrence, K.S.; Weaver, D.B.; Locy, R.D. Transcriptome Analysis of Cotton (Gossypium hirsutum L.) Genotypes that are Susceptible, Resistant, and Hypersensitive to Reniform Nematode (Rotylenchulus Reniformis). PLoS ONE 2015, 10, e0143261.
- Jones, J.T.; Haegeman, A.; Danchin, E.G.J.; Gaur, H.S.; Helder, J.; Jones, M.G.K.; Kikuchi, T.; Manzanilla-Lopez, R.; Palomares-Rius, J.E.; Wesemael, W.M.L.; et al. Top 10 Plant-Parasitic Nematodes in Molecular Plant Pathology. Mol. Plant Pathol. 2013, 14, 946–961.
- Rybicki, E.P. A Top Ten List for Economically Important Plant Viruses. Arch. Virol. 2015, 160, 17–20.
- Gorovits, R.; Moshe, A.; Amrani, L.; Kleinberger, R.; Anfoka, G.; Czosnek, H. The Six Tomato Yellow Leaf Curl Virus Genes Expressed Individually in Tomato Induce Different Levels of Plant Stress Response Attenuation. Cell Stress Chaperones 2017, 22, 345–355.
- Cheng, L.; Zou, Y.; Ding, S.; Zhang, J.; Yu, X.; Cao, J.; Lu, G. Polyamine Accumulation in Transgenic Tomato Enhances the Tolerance to High Temperature Stress. J. Integr. Plant Biol. 2009, 51, 489–499.
- Srivastava, S.; Pathak, A.D.; Gupta, P.S.; Shrivastava, A.K.; Srivastava, A.K. Hydrogen Peroxide-Scavenging Enzymes Impart Tolerance to High Temperature Induced Oxidative Stress in Sugarcane. J. Environ. Biol. 2012, 33, 657.
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat Tolerance in Plants: An Overview. Environ. Exp. Bot. 2007, 61, 199–223.
- Rodriguez, M.; Canales, E.; Borras-Hidalgo, O. Molecular Aspects of Abiotic Stress in Plants. Biotecnol. Apl. 2005, 22, 1–10.
- Tan, W.; Wei Meng, Q.; Brestic, M.; Olsovska, K.; Yang, X. Photosynthesis is Improved by Exogenous Calcium in Heat-Stressed Tobacco Plants. J. Plant Physiol. 2011, 168, 2063–2071.
- Jiang, C.; Xu, J.; Zhang, H.A.O.; Zhang, X.; Shi, J.; Li, M.I.N.; Ming, F. A Cytosolic Class I Small Heat Shock Protein, RcHSP17. 8, of Rosa Chinensis Confers Resistance to a Variety of Stresses to Escherichia coli, Yeast and Arabidopsis thaliana. Plant Cell Environ. 2009, 32, 1046–1059.
- Demirevska-Kepova, K.; Holzer, R.; Simova-Stoilova, L.; Feller, U. Heat Stress Effects on Ribulose-1, 5-Bisphosphate Carboxylase/Oxygenase, Rubisco Binding Protein and Rubisco Activase in Wheat Leaves. Biol. Plant 2005, 49, 521–525.
- Djanaguiraman, M.; Annie Sheeba, J.; Durga Devi, D.; Bangarusamy, U. Cotton Leaf Senescence can be Delayed by Nitrophenolate Spray Through Enhanced Antioxidant Defence System. J. Agron. Crop. Sci. 2009, 195, 213–224.
- Young, L.W.; Wilen, R.W.; Bonham-Smith, P.C. High Temperature Stress of Brassica napus During Flowering Reduces Micro-And Megagametophyte Fertility, Induces Fruit Abortion, and Disrupts Seed Production. J. Exp. Bot. 2004, 55, 485–495.
- Tubiello, F.N.; Soussana, J.F.; Howden, S.M. Crop and Pasture Response to Climate Change. Proc. Natl. Acad. Sci. USA 2007, 104, 19686–19690.
- Wang, X.; Cai, J.; Liu, F.; Jin, M.; Yu, H.; Jiang, D.; Wollenweber, B.; Dai, T.; Cao, W. Pre-Anthesis High Temperature Acclimation Alleviates the Negative Effects of Post-Anthesis Heat Stress on Stem Stored Carbohydrates Remobilization and Grain Starch Accumulation in Wheat. J. Cereal Sci. 2012, 55, 331–336.
- Karuppanapandian, T.; Wang, H.W.; Prabakaran, N.; Jeyalakshmi, K.; Kwon, M.; Manoharan, K.; Kim, W. 2, 4-Dichlorophenoxyacetic Acid-Induced Leaf Senescence in Mung Bean (Vigna radiata L. Wilczek) and Senescence Inhibition by Co-Treatment with Silver Nanoparticles. Plant Physiol. Biochem. 2011, 49, 168–177.
- Moller, I.M.; Jensen, P.E.; Hansson, A. Oxidative Modifications to Cellular Components in Plants. Annu. Rev. Plant Biol. 2007, 58, 459–481.
- Zhang, J.; Jia, W.; Yang, J.; Ismail, A.M. Role of ABA in Integrating Plant Responses to Drought and Salt Stresses. Field Crop. Res. 2006, 97, 111–119.
- Banon, S.; Fernandez, J.A.; Franco, J.A.; Torrecillas, A.; Alarcon, J.J.; Sanchez-Blanco, M.J. Effects of Water Stress and Night Temperature Preconditioning on Water Relations and Morphological and Anatomical Changes of Lotus Creticus Plants. Sci. Hortic. (Amst) 2004, 101, 333–342.
- Sato, S.; Kamiyama, M.; Iwata, T.; Makita, N.; Furukawa, H.; Ikeda, H. Moderate Increase of Mean Daily Temperature Adversely Affects Fruit Set of Lycopersicon esculentum by Disrupting Specific Physiological Processes in Male Reproductive Development. Ann. Bot. 2006, 97, 731–738.
- Morales, D.; Rodriguez, P.; Dell’Amico, J.; Nicolas, E.; Torrecillas, A.; Sanchez-Blanco, M.J. High-Temperature Preconditioning and Thermal Shock Imposition Affects Water Relations, Gas Exchange and Root Hydraulic Conductivity in Tomato. Biol. Plant 2003, 47, 203.
- Wahid, A.; Close, T.J. Expression of Dehydrins Under Heat Stress and Their Relationship with Water Relations of Sugarcane Leaves. Biol. Plant 2007, 51, 104–109.
- Salvucci, M.E.; Crafts-Brandner, S.J. Inhibition of Photosynthesis by Heat Stress: The Activation State of Rubisco as a Limiting Factor in Photosynthesis. Physiol. Plant 2004, 120, 179–186.
- Wahid, A.; Ghazanfar, A. Possible Involvement of Some Secondary Metabolites in Salt Tolerance of Sugarcane. J. Plant Physiol. 2006, 163, 723–730.
- Wang, L.J.; Li, S.H. Salicylic Acid-Induced Heat or Cold Tolerance in Relation to Ca2+ Homeostasis and Antioxidant Systems in Young Grape Plants. Plant Sci. 2006, 170, 685–694.
- Sharkey, T.D. Effects of Moderate Heat Stress on Photosynthesis: Importance of Thylakoid Reactions, Rubisco Deactivation, Reactive Oxygen Species, and Thermotolerance Provided by Isoprene. Plant Cell Environ. 2005, 28, 269–277.
- Laloum, T.; Martin, G.; Duque, P. Alternative Splicing Control of Abiotic Stress Responses. Trends Plant Sci. 2018, 23, 140–150.
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. Plant Microrna: A Small Regulatory Molecule with Big Impact. Dev. Biol. 2006, 289, 3–16.
- Ritosa, F. A New Puffing Pattern Induced by Heat Shock and DNP in Drosophi1ae. Cell. Mol. Life Sci. 1962, 12, 571–573.
- Tissieres, A.; Mitchell, H.K.; Tracy, U.M. Protein Synthesis in Salivary Glands of Drosophila melanogaster: Relation to Chromosome Puffs. J. Mol. Biol. 1974, 84, 389–398.
- McAlister, L.; Finkelstein, D.B. Heat Shock Proteins and Thermal Resistance in Yeast. Biochem. Biophys. Res. Commun. 1980, 93, 819–824.
- Yamamori, T.; Yura, T. Genetic Control of Heat-Shock Protein Synthesis and its Bearing on Growth and Thermal Resistance in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 1982, 79, 860–864.
- Lin, C.Y.; Roberts, J.K.; Key, J.L. Acquisition of Thermotolerance in Soybean Seedlings: Synthesis and Accumulation of Heat Shock Proteins and Their Cellular Localization. Plant Physiol. 1984, 74, 152–160.
- Bharti, K.; Nover, L. Heat Stress Response in Plants: A Complex Game with Chaperones and more than Twenty Heat Stress Transcription Factors. J. Biosci. 2004, 29, 471–487.
- Kotak, S.; Larkindale, J.; Lee, U.; Von Koskull-Doring, P.; Vierling, E.; Scharf, K.D. Complexity of the Heat Stress Response in Plants. Curr. Opin. Plant Biol. 2007, 10, 310–316.
- Sterck, L.; Rombauts, S.; Vandepoele, K.; Rouze, P.; Van De Peer, Y. How Many Genes are There in Plants (… and Why are They There)? Curr. Opin. Plant Biol. 2007, 10, 199–203.
- Lee, U.; Rioflorido, I.; Hong, S.; Larkindale, J.; Waters, E.R.; Vierling, E. The Arabidopsis ClpB/Hsp100 Family of Proteins: Chaperones for Stress and Chloroplast Development. Plant J. 2007, 49, 115–127.
- Schoffl, F.; Prandl, R.; Reindl, A. Molecular Responses to Heat Stress; Scientific Research: Wuhan, China, 1999; pp. 81–98. ISBN 1570595631.
- Schlesinger, M.J. Heat Shock Proteins. J. Biol. Chem. 1990, 265, 12111–12114.
- Liu, D.; Zhang, X.; Cheng, Y.; Takano, T.; Liu, S. RHSP90 Gene Expression in Response to Several Environmental Stresses in Rice (Oryza sativa L.). Plant Physiol. Biochem. 2006, 44, 380–386.
- Waters, E.R.; Lee, G.J.; Vierling, E. Evolution, Structure and Function of the Small Heat Shock Proteins in Plants. J. Exp. Bot. 1996, 47, 325–338.
- Korotaeva, N.E.; Antipina, A.I.; Grabelnykh, O.I.; Varakina, N.N.; Borovskii, G.B.; Voinikov, V.K. Mitochondrial Low-Molecular-Weight Heat-Shock Proteins and the Tolerance of Cereal Mitochondria to Hyperthermia. Russ. J. Plant Physiol. 2001, 48, 798–803.
- Nieto-Sotelo, J.; Martinez, L.M.; Ponce, G.; Cassab, G.I.; Alagon, A.; Meeley, R.B.; Ribaut, J.M.; Yang, R. Maize HSP101 Plays Important Roles in Both Induced and Basal Thermotolerance and Primary Root Growth. Plant Cell 2002, 14, 1621–1633.
- Neumann, D.; Emmermann, M.; Thierfelder, J.M.; Zur Nieden, U.; Clericus, M.; Braun, H.P.; Nover, L.; Schmitz, U.K. HSP68—A DnaK-Like Heat-Stress Protein of Plant Mitochondria. Planta 1993, 190, 32–43.
- Young, T.E.; Ling, J.; Geisler-Lee, C.J.; Tanguay, R.L.; Caldwell, C.; Gallie, D.R. Developmental and Thermal Regulation of the Maize Heat Shock Protein, HSP101. Plant Physiol. 2001, 127, 777–791.
- Heckathorn, S.A.; Downs, C.A.; Coleman, J.S. Nuclear-Encoded Chloroplast Proteins Accumulate in the Cytosol During Severe Heat Stress. Int. J. Plant Sci. 1998, 159, 39–45.
- Frova, C.; Taramino, G.; Binelli, G. Heat-Shock Proteins During Pollen Development in Maize. Dev. Genet. 1989, 10, 324–332.
- Ristic, Z.; Williams, G.; Yang, G.; Martin, B.; Fullerton, S. Dehydration, Damage to Cellular Membranes, and Heat-Shock Proteins in Maize Hybrids From Different Climates. J. Plant Physiol. 1996, 149, 424–432.
- Mittler, R.; Kim, Y.; Song, L.; Coutu, J.; Coutu, A.; Ciftci-Yilmaz, S.; Lee, H.; Stevenson, B.; Zhu, J.K. Gain-And Loss-Of-Function Mutations in Zat10 Enhance the Tolerance of Plants to Abiotic Stress. FEBS Lett. 2006, 580, 6537–6542.
- Wahid, A. Physiological Implications of Metabolite Biosynthesis for Net Assimilation and Heat-Stress Tolerance of Sugarcane (Saccharum officinarum) Sprouts. J. Plant Res. 2007, 120, 219–228.
- Singla, J.; Krattinger, S.G. Biotic Stress Resistance Genes in Wheat; Elsevier: Amsterdam, The Netherlands, 2016; ISBN 978-0-08-100596-5.
- Whitham, S.A.; Quan, S.; Chang, H.; Cooper, B.; Estes, B.; Zhu, T.; Wang, X.; Hou, Y. Diverse RNA Viruses Elicit the Expression of Common Sets of Genes in Susceptible Arabidopsis Thaliana Plants. Plant J. 2003, 33, 271–283.
- Gorovits, R.; Moshe, A.; Ghanim, M.; Czosnek, H. Recruitment of the Host Plant Heat Shock Protein 70 by Tomato Yellow Leaf Curl Virus Coat Protein is Required for Virus Infection. PLoS ONE 2013, 8, e70280.
- Gorovits, R.; Czosnek, H. The Involvement of Heat Shock Proteins in the Establishment of Tomato Yellow Leaf Curl Virus Infection. Front. Plant Sci. 2017, 8, 355.
- Alam, S.B.; Rochon, D. Cucumber Necrosis Virus Recruits Cellular Heat Shock Protein 70 Homologs at Several Stages of Infection. J. Virol. 2016, 90, 3302–3317.
- Chen, Z.; Zhou, T.; Wu, X.; Hong, Y.; Fan, Z.; Li, H. Influence of Cytoplasmic Heat Shock Protein 70 on Viral Infection of Nicotiana benthamiana. Mol. Plant Pathol. 2008, 9, 809–817.
- Mine, A.; Hyodo, K.; Tajima, Y.; Kusumanegara, K.; Taniguchi, T.; Kaido, M.; Mise, K.; Taniguchi, H.; Okuno, T. Differential Roles of Hsp70 and Hsp90 in the Assembly of the Replicase Complex of a Positive-Strand RNA Plant Virus. J. Virol. 2012, 86, 91–104.
- Senthil, G.; Liu, H.; Puram, V.G.; Clark, A.; Stromberg, A.; Goodin, M.M. Specific and Common Changes in Nicotiana benthamiana Gene Expression in Response to Infection by Enveloped Viruses. J. Gen. Virol. 2005, 86, 2615–2625.
- Yu, L.; Wang, W.; Zeng, S.; Chen, Z.; Yang, A.; Shi, J.; Zhao, X.; Song, B. Label-Free Quantitative Proteomics Analysis of Cytosinpeptidemycin Responses in Southern Rice Black-Streaked Dwarf Virus-Infected Rice. Pestic. Biochem. Physiol. 2018, 147, 20–26.
- Jiang, S.; Lu, Y.; Li, K.; Lin, L.; Zheng, H.; Yan, F.; Chen, J. Heat Shock Protein 70 is Necessary for R ice Stripe Virus Infection in Plants. Mol. Plant Pathol. 2014, 15, 907–917.
- Makarova, S.; Makhotenko, A.; Spechenkova, N.; Love, A.J.; Kalinina, N.O.; Taliansky, M. Interactiver Responses of Potato (Solanum tuberosum L.) Plants to Heat Stress and Infection with Potato Virus Y. Front. Microbiol. 2018, 9, 2582.
- Maimbo, M.; Ohnishi, K.; Hikichi, Y.; Yoshioka, H.; Kiba, A. Induction of a Small Heat Shock Protein and its Functional Roles in Nicotiana Plants in the Defense Response Against Ralstonia solanacearum. Plant Physiol. 2007, 145, 1588–1599.
- Ito, M.; Ohnishi, K.; Hikichi, Y.; Kiba, A. Molecular Chaperons and Co-Chaperons, Hsp90, RAR1, and SGT1 Negatively Regulate Bacterial wilt Disease Caused by Ralstonia solanacearum in Nicotiana benthamiana. Plant Signal. Behav. 2014, 10, e970410.
- Garofalo, C.G.; Garavaglia, B.S.; Dunger, G.; Gottig, N.; Orellano, E.G.; Ottado, J. Expression Analysis of Small Heat Shock Proteins During Compatible and Incompatible Plant-Pathogen Interactions. Adv. Stud. Biol. 2009, 5, 197–205.
- Bricchi, I.; Bertea, C.M.; Occhipinti, A.; Paponov, I.A.; Maffei, M.E. Dynamics of Membrane Potential Variation and Gene Expression Induced by Spodoptera littoralis, Myzus persicae, and pseudomonas syringae in Arabidopsis. PLoS ONE 2012, 7, e46673.
- Pavlova, E.L.; Rikhvanov, E.G.; Tauson, E.L.; Varakina, N.N.; Gamburg, K.Z.; Rusaleva, T.M.; Borovskii, G.B.; Voinikov, V.K. Effect of Salicylic Acid on the Development of Induced Thermotolerance and Induction of Heat Shock Protein Synthesis in the Arabidopsis Thaliana Cell Culture. Russ. J. Plant Physiol. 2009, 56, 68–73.
- Sarkar, N.K.; Kim, Y.K.; Grover, A. Rice sHsp Genes: Genomic Organization and Expression Profiling Under Stress and Development. BMC Genom. 2009, 10, 393.
- Van Ooijen, G.; Lukasik, E.; Van Den Burg, H.A.; Vossen, J.H.; Cornelissen, B.J.C.; Takken, F.L.W. The Small Heat Shock Protein 20 RSI2 Interacts with and is Required for Stability and Function of Tomato Resistance Protein I-2. Plant J. 2010, 63, 563–572.
- Pan, X.; Zhu, B.; Luo, Y.; Fu, D. Unraveling the Protein Network of Tomato Fruit in Response to Necrotrophic Phytopathogenic Rhizopus nigricans. PLoS ONE 2013, 8, e73034.
- Piterkova, J.; Luhova, L.; Mieslerova, B.; Lebeda, A.; Petrivalsky, M. Nitric Oxide and Reactive Oxygen Species Regulate the Accumulation of Heat Shock Proteins in Tomato Leaves in Response to Heat Shock and Pathogen Infection. Plant Sci. 2013, 207, 57–65.
- Husselmann, L.H.H. Analysis of the Early Events in the Interaction between Venturia inaequalis and the Susceptible Golden Delicious Apple (Malus domestica Borkh.). Ph.D. Thesis, University of the Western Cape, Belleville, Cape Town, Western Cape, South Africa, 2014.
- Yogendra, K.N.; Kumar, A.; Sarkar, K.; Li, Y.; Pushpa, D.; Mosa, K.A.; Duggavathi, R.; Kushalappa, A.C. Transcription Factor StWRKY1 Regulates Phenylpropanoid Metabolites Conferring Late Blight Resistance in Potato. J. Exp. Bot. 2015, 66, 7377–7389.
- Kallamadi, P.R.; Dandu, K.; Kirti, P.B. An Insight into Powdery Mildew–Infected, Susceptible, Resistant and Immune Sunflower Genotypes. Proteomics 2018, 18, 1700418.
- Calderwood, S.K. Heat Shock Proteins and Plants; Springer International Publishing: Cham, Switzerland; Boston, MA, USA, 2016; ISBN 9783319463391.
- Lopes-Caitar, V.S.; De Carvalho, M.C.C.G.; Darben, L.M.; Kuwahara, M.K.; Nepomuceno, A.L.; Dias, W.P.; Abdelnoor, R.V.; Marcelino-Guimaraes, F.C. Genome-Wide Analysis of the Hsp 20 Gene Family in Soybean: Comprehensive Sequence, Genomic Organization and Expression Profile Analysis Under Abiotic and Biotic Stresses. BMC Genom. 2013, 14, 577.
- Fuganti, R.; Machado, M.D.F.P.D.; Lopes, V.S.; Silva, J.F.V.; Arias, C.A.A.; Marin, S.R.R.; Binneck, E.; Abdelnoor, R.V.; Marcelino, F.C.; Nepomuceno, A.L. Size of A T (n) Insertions in Promoter Region Modulates Gmhsp17.6-L mRNA Transcript Levels. Biomed. Res. Int. 2010, 2010, 847673.
- Kandoth, P.K.; Ithal, N.; Recknor, J.; Maier, T.; Nettleton, D.; Baum, T.J.; Mitchum, M.G. The Soybean Rhg1 Locus for Resistance to the Soybean Cyst Nematode Heterodera Glycines Regulates Expression of a Large Number of Stress-And Defense-Related Genes in Degenerating Feeding Cells. Plant Physiol. 2011, 155, 960–975.
- Escobar, C.; Barcala, M.; Portillo, M.; Almoguera, C.; Jordano, J.; Fenoll, C. Induction of the Hahsp17. 7G4 Promoter by Root-Knot Nematodes: Involvement of Heat-Shock Elements in Promoter Activity in Giant Cells. Mol. Plant Microbe Interact. 2003, 16, 1062–1068.
- Barcala, M.; Garcia, A.; Cubas, P.; Almoguera, C.; Jordano, J.; Fenoll, C.; Escobar, C. Distinct Heat-Shock Element Arrangements that Mediate the Heat Shock, But Not the Late-Embryogenesis Induction of Small Heat-Shock Proteins, Correlate with Promoter Activation in Root-Knot Nematode Feeding Cells. Plant Mol. Biol. 2008, 66, 151–164.
- Bhattarai, K.K.; Li, Q.; Liu, Y.; Dinesh-Kumar, S.P.; Kaloshian, I. The MI-1-Mediated Pest Resistance Requires Hsp90 and Sgt1. Plant Physiol. 2007, 144, 312–323.
- Lourenço-Tessutti, I.; Antonino De Souza, J., Jr.; Martins-De-SA, D.; Viana, A.A.; Carneiro, R.; Togawa, R.; Engler, J.; Batista, J.; Cristina Mattar Silva, M.; Fragoso, R.; et al. Knock-Down of Heat-Shock Protein 90 and Isocitrate Lyase gene expression reduced Root-Knot Nematode reproduction. Phytopathology 2015, 105, 628–637.
- Ahuja, I.; De Vos, R.C.H.; Bones, A.M.; Hall, R.D. Plant Molecular Stress Responses Face Climate Change. Trends Plant Sci. 2010, 15, 664–674.
- Zhu, J.K. Plant Salt Tolerance. Trends Plant Sci. 2001, 6, 66–71.
- Reguera, M.; Peleg, Z.; Blumwald, E. Targeting Metabolic Pathways for Genetic Engineering Abiotic Stress-Tolerance in Crops. Biochim. Biophys. Acta. Gene Regul. Mech. 2012, 1819, 186–194.
- Id, H.W.; Shi, N.; An, X.; Liu, C.; Fu, H.; Cao, L.; Feng, Y. Candidate Genes for Yellow Leaf Color in Common Wheat (Triticum aestivum L.) and Major Related Metabolic Pathways According to Transcriptome Profiling. Int. J. Mol. Sci. 2018, 19, 1594.
- Chauhan, H.; Khurana, N.; Nijhavan, A.; Khurana, J.P.; Khurana, P. The Wheat Chloroplastic Small Heat Shock Protein (sHSP26) is Involved in Seed Maturation and Germination and Imparts Tolerance to Heat Stress. Plant Cell Environ. 2012, 35, 1912–1931.
- Singla, S.L.; Pareek, A.; Grover, A. Plant Hsp100 Family with Special Reference to Rice. J. Biosci. 1998, 23, 337–345.
- Prasad, B.D.; Goel, S.; Krishna, P. In Silico Identification of Carboxylate Clamp Type Tetratricopeptide Repeat Proteins in Arabidopsis and Rice as Putative Co-Chaperones of Hsp90/Hsp70. PLoS ONE 2010, 5, e12761.
- Xu, J.; Xue, C.; Xue, D.; Zhao, J.; Gai, J.; Guo, N.; Xing, H. Overexpression of GmHsp90s, a Heat Shock Protein 90 (Hsp90) Gene Family Cloning From Soybean, Decrease Damage of Abiotic Stresses in Arabidopsis Thaliana. PLoS ONE 2013, 8, e69810.
- Pratt, W.B.; Toft, D.O. Regulation of Signaling Protein Function and Trafficking by the hsp90/hsp70-Based Chaperone Machinery. Exp. Biol. Med. 2003, 228, 111–133.
- Dupuis, I.; Dumas, C. Influence of Temperature Stress on in Vitro Fertilization and Heat Shock Protein Synthesis in Maize (Zea mays L.) Reproductive Tissues. Plant Physiol. 1990, 94, 665–670.
- Bolhassani, A.; Agi, E. Heat shock proteins in infection. Clin. Chim. Acta 2019, 498, 90–100.
- Bita, C.; Gerats, T. Plant Tolerance to High Temperature in a Changing Environment: Scientific Fundamentals and Production of Heat Stress-Tolerant Crops. Front. Plant Sci. 2013, 4, 273.
- Yamada, K.; Fukao, Y.; Hayashi, M.; Fukazawa, M.; Suzuki, I.; Nishimura, M. Cytosolic HSP90 Regulates the Heat Shock Response that is Responsible for Heat Acclimation in Arabidopsis Thaliana. J. Biol. Chem. 2007, 282, 37794–37804.
- Xu, X.; Song, H.; Zhou, Z.; Shi, N.; Ying, Q.; Wang, H. Functional Characterization of AtHsp90.3 in Saccharomyces cerevisiae and Arabidopsis Thaliana Under Heat Stress. Biotechnol. Lett. 2010, 32, 979–987.
- Kim, S.R.; An, G. Rice Chloroplast-Localized Heat Shock Protein 70, OsHsp70CP1, is Essential for Chloroplast Development Under High-Temperature Conditions. J. Plant Physiol. 2013, 170, 854–863.
- Jungkunz, I.; Link, K.; Vogel, F.; Voll, L.M.; Sonnewald, S.; Sonnewald, U. AtHsp70-15-Deficient Arabidopsis Plants are Characterized by Reduced Growth, a Constitutive Cytosolic Protein Response and Enhanced Resistance to TuMV. Plant J. 2011, 66, 983–995.
- Liu, J.; Pang, X.; Cheng, Y.; Yin, Y.; Zhang, Q.; Su, W.; Hu, B. The Hsp70 Gene Family in Solanum tuberosum: Genome-Wide Identification, Phylogeny, and Expression Patterns. Sci. Rep. 2018, 8, 16628.
- Duck, N.B.; Folk, W.R. Hsp70 Heat Shock Protein Cognate is Expressed and Stored in Developing Tomato Pollen. Plant Mol. Biol. 1994, 26, 1031–1039.
- Yu, J.; Cheng, Y.; Feng, K.; Ruan, M.; Ye, Q.; Wang, R.; Li, Z.; Zhou, G.; Yao, Z.; Yang, Y. Genome-Wide Identification and Expression Profiling of Tomato Hsp20 Gene Family in Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2016, 7, 1215.
- Derocher, A.E.; Helm, K.W.; Lauzon, L.M.; Vierling, E. Expression of a Conserved Family of Cytoplasmic Low Molecular Weight Heat Shock Proteins during Heat Stress and Recovery. Plant Physiol. 1991, 96, 1038–1047.
- Zhang, S.; Wang, S.; Lv, J.; Liu, Z.; Wang, Y.; Ma, N.; Meng, Q. SUMO E3 Ligase SlSIZ1 Facilitates Heat Tolerance in Tomato. Plant Cell Physiol. 2017, 59, 58–71.
- Guo, M.; Liu, J.; Ma, X.; Zhai, Y.; Gong, Z.; Lu, M. Plant Science Genome-Wide Analysis of the Hsp70 Family Genes in Pepper (Capsicum annuum L.) and Functional Identification of CaHsp70-2 Involvement in Heat Stress. Plant Sci. 2016, 252, 246–256.
- Usman, M.G.; Rafii, M.Y.; Martini, M.Y.; Yusuff, O.A. Introgression of Heat Shock Protein (Hsp70 and sHsp) Genes into the Malaysian Elite Chilli Variety Kulai (Capsicum annuum L.) Through the Application of Marker-Assisted Backcrossing (MAB). Cell Stress Chaperones 2018, 23, 223–234.
- Haq, S.U.L.; Khan, A.; Ali, M.; Gai, W.X.; Zhang, H.X.; Yu, Q.H.; Yang, S.B.; Wei, A.M.; Gong, Z.H. Knockdown of CaHSP60-6 Confers Enhanced Sensitivity to Heat Stress in Pepper (Capsicum annuum L.). Planta 2019.
- Guo, M.; Liu, J.H.; Lu, J.P.; Zhai, Y.F.; Wang, H.; Gong, Z.H.; Wang, S.B.; Lu, M.H. Genome-Wide Analysis of the CaHsp20 Gene Family in Pepper: Comprehensive Sequence and Expression Profile Analysis Under Heat Stress. Front. Plant Sci. 2015, 6, 806.
- Huang, L.; Cheng, G.; Khan, A.; Wei, A.; Yu, Q.; Yang, S.; Luo, D. CaHSP16.4, a Small Heat Shock Protein Gene in Pepper, is Involved in Heat and Drought Tolerance. Protoplasma 2019, 256, 39–51.
- Lee, S.S.; Jung, W.Y.; Park, H.J.; Lee, A.; Kwon, S.; Cho, H.S. Genome-Wide Analysis of Alternative Splicing in an Inbred Cabbage (Brassica oleracea L.) line‘HO’in Response to Heat Stress. Curr. Genom. 2018, 19, 12–20.
- Chen, J.; Gao, T.; Wan, S.; Zhang, Y.; Yang, J.; Yu, Y.; Wang, W. Genome-Wide Identification, Classification and Expression Analysis of the HSP Gene Superfamily in Tea Plant (Camellia sinensis). Int. J. Mol. Sci. 2018, 19, 2633.
- Song, G.; Yuan, S.; Wen, X.; Xie, Z.; Lou, L.; Hu, B.; Cai, Q.; Xu, B. Transcriptome Analysis of Cd-Treated Switchgrass Root Revealed Novel Transcripts and the Importance of HSF / HSP Network in Switchgrass Cd Tolerance. Plant Cell Rep. 2018, 37, 1485–1497.
- Li, Z.; Long, R.; Zhang, T.; Wang, Z.; Zhang, F.; Yang, Q.; Kang, J.; Sun, Y. Molecular Cloning and Functional Analysis of the Drought Tolerance Gene MsHSP70 From Alfalfa (Medicago sativa L.). J. Plant Res. 2017, 130, 387–396.
- Singh, R.K.; Jaishankar, J.; Muthamilarasan, M.; Shweta, S.; Dangi, A.; Prasad, M. Genome-Wide Analysis of Heat Shock Proteins in C 4 Model, Foxtail Millet Identifies Potential Candidates for Crop Improvement Under Abiotic Stress. Sci. Rep. 2016, 6, 32641.
- Zhang, Y.; Sun, M.; Zhang, Q. Proteomic Analysis of the Heat Stress Response in Leaves of Two Contrasting Chrysanthemum Varieties. Plant Omics 2014, 7, 229.
- Bae, M.S.; Cho, E.J.; Choi, E.; Park, O.K. Analysis of the Arabidopsis Nuclear Proteome and its Response to Cold Stress. Plant J. 2003, 36, 652–663.
- Kumar, N.; Suyal, D.C.; Sharma, I.P.; Verma, A.; Singh, H. Elucidating Stress Proteins in Rice (Oryza sativa L.) Genotype Under Elevated Temperature: A Proteomic Approach to Understand Heat Stress Response. 3 Biotech 2017, 7, 205.
- Wang, X.; Zhang, H.; Shao, L.Y.; Yan, X.; Peng, H.; Ouyang, J.X.; Li, S.B. Expression and Function Analysis of a Rice OsHSP40 Gene Under Salt Stress. Genes Genom. 2018, 11, 1–8.
- Kollipara, K.P.; Saab, I.N.; Wych, R.D.; Lauer, M.J.; Singletary, G.W. Expression Profiling of Reciprocal Maize Hybrids Divergent for Cold Germination and Desiccation Tolerance. Plant Physiol. 2002, 129, 974–992.
- Kosova, K.; Vitamvas, P.; Planchon, S.; Renaut, J.; Vankova, R.; Prasil, I.T. Proteome Analysis of Cold Response in Spring and Winter Wheat (Triticum aestivum) Crowns Reveals Similarities in Stress Adaptation and Differences in Regulatory Processes between the Growth Habits. J. Proteome Res. 2013, 12, 4830–4845.
- Vitamvas, P.; Prasil, I.T.; Kosova, K.; Planchon, S.; Renaut, J. Analysis of Proteome and Frost Tolerance in Chromosome 5A and 5B Reciprocal Substitution Lines Between Two Winter Wheats During Long-Term Cold Acclimation. Proteomics 2012, 12, 68–85.
- Hlavackova, I.; Vitamvas, P.; Santrueek, J.; Kosova, K.; Zelenkova, S.; Prasil, I.T.; Ovesna, J.; Hynek, R.; Kodicek, M. Proteins Involved in Distinct Phases of Cold Hardening Process in Frost Resistant Winter Barley (Hordeum vulgare L.) Cv Luxor. Int. J. Mol. Sci. 2013, 14, 8000–8024.
- Cui, S.; Huang, F.; Wang, J.; Ma, X.; Cheng, Y.; Liu, J. A Proteomic Analysis of Cold Stress Responses in Rice Seedlings. Proteomics 2005, 5, 3162–3172.
- Suzuki, K.; Aoki, N.; Matsumura, H.; Okamura, M.; Ohsugi, R.; Shimono, H. Cooling Water Before Panicle Initiation Increases Chilling-Induced Male Sterility and Disables Chilling-Induced Expression of Genes Encoding OsFKBP65 and Heat Shock Proteins in Rice Spikelets. Plant Cell Environ. 2015, 38, 1255–1274.
- Degand, H.; Faber, A.; Dauchot, N.; Mingeot, D.; Watillon, B.; Van Cutsem, P.; Morsomme, P.; Boutry, M. Proteomic Analysis of Chicory Root Identifies Proteins Typically Involved in Cold Acclimation. Proteomics 2009, 9, 2903–2907.
- Reddy, R.K.; Chaudhary, S.; Patil, P.; Krishna, P. The 90 kDa Heat Shock Protein (hsp90) is Expressed Throughout Brassica napus Seed Development and Germination. Plant Sci. 1998, 131, 131–137.
- Renaut, J.; Lutts, S.; Hoffmann, L.; Hausman, J. Responses of Poplar to Chilling Temperatures: Proteomic and Physiological Aspects. Plant Biol. 2004, 6, 81–90.
- Taylor, N.L.; Heazlewood, J.L.; Day, D.A.; Millar, A.H. Differential Impact of Environmental Stresses on the Pea Mitochondrial Proteome. Mol. Cell. Proteom. 2005, 4, 1122–1133.
- Balbuena, T.S.; Salas, J.J.; Martinez-Force, E.; Garces, R.; Thelen, J.J. Proteome Analysis of Cold Acclimation in Sunflower. J. Proteome Res. 2011, 10, 2330–2346.
- Sabehat, A.; Lurie, S.; Weiss, D. Expression of Small Heat-Shock Proteins at Low Temperatures: A Possible Role in Protecting Against Chilling Injuries. Plant Physiol. 1998, 117, 651–658.
- Sun, J.; Chen, J.; Kuang, J.; Chen, W.; Lu, W. Expression of sHSP Genes as Affected by Heat Shock and Cold Acclimation in Relation to Chilling Tolerance in Plum Fruit. Postharvest Biol. Technol. 2010, 55, 91–96.
- Rozenzvieg, D.; Elmaci, C.; Samach, A.; Lurie, S.; Porat, R. Isolation of Four Heat Shock Protein cDNAs From Grapefruit Peel Tissue and Characterization of Their Expression in Response to Heat and Chilling Temperature Stresses. Physiol. Plant 2004, 121, 421–428.
- Sobhanian, H.; Aghaei, K.; Komatsu, S. Changes in the Plant Proteome Resulting From Salt Stress: Toward the Creation of Salt-Tolerant Crops? J. Proteom. 2011, 74, 1323–1337.
- Wang, M.; Peng, Z.; Li, C.; Li, F.; Liu, C.; Xia, G. Proteomic Analysis on a High Salt Tolerance Introgression Strain of Triticum aestivum/Thinopyrum ponticum. Proteomics 2008, 8, 1470–1489.
- Ngara, R.; Ndimba, B.K. Understanding the Complex Nature of Salinity and Drought-Stress Response in Cereals Using Proteomics Technologies. Proteomics 2014, 14, 611–621.
- Han, F.; Chen, H.; Li, X.J.; Yang, M.F.; Liu, G.S.; Shen, S.H. A Comparative Proteomic Analysis of Rice Seedlings Under Various High-Temperature stresses. Biochim. Biophys. Acta. Proteins Proteom. 2009, 1794, 1625–1634.
- Muthusamy, S.K.; Dalal, M.; Chinnusamy, V.; Bansal, K.C. Differential Regulation of Genes Coding for Organelle and Cytosolic ClpATPases Under Biotic and Abiotic Stresses in Wheat. Front. Plant Sci. 2016, 7, 929.
- Mishra, R.C.; Grover, A. Constitutive Over-Expression of Rice ClpD1 Protein Enhances Tolerance to Salt and Desiccation Stresses in Transgenic Arabidopsis Plants. Plant Sci. 2016, 250, 69–78.
- Song, H.; Fan, P.; Li, Y. Overexpression of Organellar and Cytosolic AtHSP90 in Arabidopsis Thaliana Impairs Plant Tolerance to Oxidative Stress. Plant Mol. Biol. Report. 2009, 27, 342–349.
- Pi, E.; Qu, L.; Hu, J.; Huang, Y.; Qiu, L.; Lu, H.; Jiang, B.; Liu, C.; Peng, T.; Zhao, Y. Mechanisms of Soybean Roots’ Tolerances to Salinity Revealed by Proteomic and Phosphoproteomic Comparisons Between Two Cultivars. Mol. Cell. Proteom. 2016, 15, 266–288.
- Manaa, A.; Ben Ahmed, H.; Valot, B.; Bouchet, J.P.; Aschi-Smiti, S.; Causse, M.; Faurobert, M. Salt and Genotype Impact on Plant Physiology and Root Proteome Variations in Tomato. J. Exp. Bot. 2011, 62, 2797–2813.
- Yer, E.N.; Baloglu, M.C.; Ayan, S. Identification and Expression Profiling of All Hsp Family Member Genes Under Salinity Stress in Different Poplar Clones. Gene 2018, 678, 324–336.
- Song, A.; Zhu, X.; Chen, F.; Gao, H.; Jiang, J.; Chen, S. A Chrysanthemum Heat Shock Protein Confers Tolerance to Abiotic Stress. Int. J. Mol. Sci. 2014, 15, 5063–5078.
- Ono, K.; Hibino, T.; Kohinata, T.; Suzuki, S.; Tanaka, Y.; Nakamura, T.; Takabe, T. Overexpression of DnaK From a Halotolerant Cyanobacterium Aphanothece halophytica Enhances the High-Temperatue Tolerance of Tobacco During Germination and Early Growth. Plant Sci. 2001, 160, 455–461.
- Alvim, F.C.; Carolino, S.M.B.; Cascardo, J.C.M.; Nunes, C.C.; Martinez, C.A.; Otoni, W.C.; Fontes, E.P.B. Enhanced Accumulation of BiP in Transgenic Plants Confers Tolerance to Water Stress. Plant Physiol. 2001, 126, 1042–1054.
- Reddy, P.S.; Kishor, P.B.K.; Seiler, C.; Kuhlmann, M.; Eschen-Lippold, L.; Lee, J.; Reddy, M.K.; Sreenivasulu, N. Unraveling Regulation of the Small Heat Shock Proteins by the Heat Shock Factor HvHsfB2c in Barley: Its Implications in Drought Stress Response and Seed Development. PLoS ONE 2014, 9, e89125.
- Shu, L.; Lou, Q.; Ma, C.; Ding, W.; Zhou, J.; Wu, J.; Feng, F.; Lu, X.; Luo, L.; Xu, G. Genetic, Proteomic and Metabolic Analysis of the Regulation of Energy Storage in Rice Seedlings in Response to Drought. Proteomics 2011, 11, 4122–4138.
- Agrawal, L.; Gupta, S.; Mishra, S.K.; Pandey, G.; Kumar, S.; Chauhan, P.S.; Chakrabarty, D.; Nautiyal, C.S. Elucidation of Complex Nature of PEG Induced Drought-Stress Response in Rice Root Using Comparative Proteomics Approach. Front. Plant Sci. 2016, 7, 1466.
- Benesova, M.; Hola, D.; Fischer, L.; Jedelsky, P.L.; Hnilicka, F.; Wilhelmova, N.; Rothova, O.; Kocova, M.; Prochazkova, D.; Honnerova, J. The Physiology and Proteomics of Drought Tolerance in Maize: Early Stomatal Closure as a Cause of Lower Tolerance to Short-Term Dehydration. PLoS ONE 2012, 7, e38017.
- Sato, Y.; Yokoya, S. Enhanced Tolerance to Drought Stress in Transgenic Rice Plants Overexpressing a Small Heat-Shock Protein, sHSP17. 7. Plant Cell Rep. 2008, 27, 329–334.
- Subba, P.; Barua, P.; Kumar, R.; Datta, A.; Soni, K.K.; Chakraborty, S.; Chakraborty, N. Phosphoproteomic Dynamics of Chickpea (Cicer arietinum L.) Reveals Shared and Distinct Components of Dehydration Response. J. Proteome Res. 2013, 12, 5025–5047.
- Augustine, S.M.; Cherian, A.V.; Syamaladevi, D.P.; Subramonian, N. Erianthus arundinaceus HSP70 (EaHSP70) Acts as a Key Regulator in the Formation of Anisotropic Interdigitation in Sugarcane (Saccharum Spp. Hybrid) in Response to Drought Stress. Plant Cell Physiol. 2015, 56, 2368–2380.
- Xu, C.; Huang, B. Comparative Analysis of Drought Responsive Proteins in Kentucky Bluegrass Cultivars Contrasting in Drought Tolerance. Crop. Sci. 2010, 50, 2543–2552.
- Burke, J.J.; Hatfield, J.L.; Klein, R.R.; Mullet, J.E. Accumulation of Heat Shock Proteins in Field-Grown Cotton. Plant Physiol. 1985, 78, 394–398.
- Bonhomme, L.; Monclus, R.; Vincent, D.; Carpin, S.; Lomenech, A.; Plomion, C.; Brignolas, F.; Morabito, D. Leaf Proteome Analysis of Eight Populus euramericana Genotypes: Genetic Variation in Drought Response and in Water-Use Efficiency Involves Photosynthesis-Related Proteins. Proteomics 2009, 9, 4121–4142.
- Giacomelli, L.; Rudella, A.; Van Wijk, K.J. High Light Response of the Thylakoid Proteome in Arabidopsis wild Type and the Ascorbate-Deficient Mutant Vtc2-2. A Comparative Proteomics Study. Plant Physiol. 2006, 141, 685–701.
- Rossel, J.B.; Wilson, I.W.; Pogson, B.J. Global Changes in Gene Expression in Response to High Light in Arabidopsis. Plant Physiol. 2002, 130, 1109–1120.
- Kropat, J.; Oster, U.; Rudiger, W.; Beck, C.F. Chlorophyll Precursors are Signals of Chloroplast Origin Involved in Light Induction of Nuclear Heat-Shock Genes. Proc. Natl. Acad. Sci. USA 1997, 94, 14168–14172.
- Rodriguez-Celma, J.; Rellan-Alvarez, R.; Abadia, A.; Abadia, J.; Lopez-Millan, A.F. Changes Induced by Two Levels of Cadmium Toxicity in the 2-DE Protein Profile of Tomato Roots. J. Proteom. 2010, 73, 1694–1706.
- Ahsan, N.; Lee, S.H.; Lee, D.G.; Lee, H.; Lee, S.W.; Bahk, J.D.; Lee, B.H. Physiological and Protein Profiles Alternation of Germinating Rice Seedlings Exposed to Acute Cadmium Toxicity. C. R. Biol. 2007, 330, 735–746.
- Yang, Y.; Li, X.; Yang, S.; Zhou, Y.; Dong, C.; Ren, J.; Sun, X.; Yang, Y. Comparative Physiological and Proteomic Analysis Reveals the Leaf Response to Cadmium-Induced Stress in Poplar (Populus yunnanensis). PLoS ONE 2015, 10, e0137396.
- Czarnecka, E.; Nagao, R.T.; Key, J.L.; Gurley, W.B. Characterization of Gmhsp26-A, a Stress Gene Encoding a Divergent Heat Shock Protein of Soybean: Heavy-Metal-Induced Inhibition of Intron Processing. Mol. Cell. Biol. 1988, 8, 1113–1122.
- Kosova, K.; Vitamvas, P.; Prasil, I.T.; Renaut, J. Plant Proteome Changes Under Abiotic Stress—Contribution of Proteomics Studies to Understanding Plant Stress Response. J. Proteom. 2011, 74, 1301–1322.
- Lee, J.; Ahn, Y.J. Heterologous Expression of a Carrot Small Heat Shock Protein Increased Escherichia Coli Viability Under Lead and Arsenic Stresses. HortScience 2013, 48, 1323–1326.
- Sarry, J.; Kuhn, L.; Ducruix, C.; Lafaye, A.; Junot, C.; Hugouvieux, V.; Jourdain, A.; Bastien, O.; Fievet, J.B.; Vailhen, D. The Early Responses of Arabidopsis Thaliana Cells to Cadmium Exposure Explored by Protein and Metabolite Profiling Analyses. Proteomics 2006, 6, 2180–2198.
- Navascues, J.; Perez-Rontome, C.; Sanchez, D.H.; Staudinger, C.; Wienkoop, S.; Rellan-Alvarez, R.; Becana, M. Oxidative Stress is a Consequence, Not a Cause, of Aluminum Toxicity in the Forage Legume Lotus corniculatus. New Phytol. 2012, 193, 625–636.
- Banti, V.; Mafessoni, F.; Loreti, E.; Alpi, A.; Perata, P. The Heat-Inducible Transcription Factor HsfA2 Enhances Anoxia Tolerance in Arabidopsis. Plant Physiol. 2010, 152, 1471–1483.
- Huther, C.M.; Martinazzo, E.G.; Rombaldi, C.V.; Bacarin, M.A. Effects of Flooding Stress in Micro-Tom’tomato Plants Transformed with Different Levels of Mitochondrial sHSP23. 6. Braz. J. Biol. 2017, 77, 43–51.
- Qi, Y.; Wang, H.; Zou, Y.; Liu, C.; Liu, Y.; Wang, Y.; Zhang, W. Over-Expression of Mitochondrial Heat Shock Protein 70 Suppresses Programmed Cell Death in Rice. Febs Lett. 2011, 585, 231–239.
- Chen, Y.; Chen, X.; Wang, H.; Bao, Y.; Zhang, W. Examination of the Leaf Proteome During Flooding Stress and the Induction of Programmed Cell Death in Maize. Proteome Sci. 2014, 12, 33.
- Komatsu, S.; Makino, T.; Yasue, H. Proteomic and Biochemical Analyses of the Cotyledon and Root of Flooding-Stressed Soybean Plants. PLoS ONE 2013, 8, e65301.
More
Information
Subjects:
Agriculture, Dairy & Animal Science
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.0K
Revisions:
2 times
(View History)
Update Date:
29 Oct 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




