
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Anna Pasetto | + 2603 word(s) | 2603 | 2021-09-06 04:52:12 | | | |
| 2 | Lindsay Dong | Meta information modification | 2603 | 2021-09-28 07:47:34 | | |
Video Upload Options
Hepatocellular carcinoma (HCC) causes many deaths worldwide, and current treatments have limitations. Immunotherapies have shown the most promising clinical outcomes for advanced HCC. However, there are many patients with HCC who still respond poorly to these treatments. Circulating biomarkers that can easily be obtained through blood sampling are promising in predicting treatment responses, since they are minimally invasive and enable us to constantly monitor disease progression.
1. Introduction
2. Liquid Biopsy in HCC
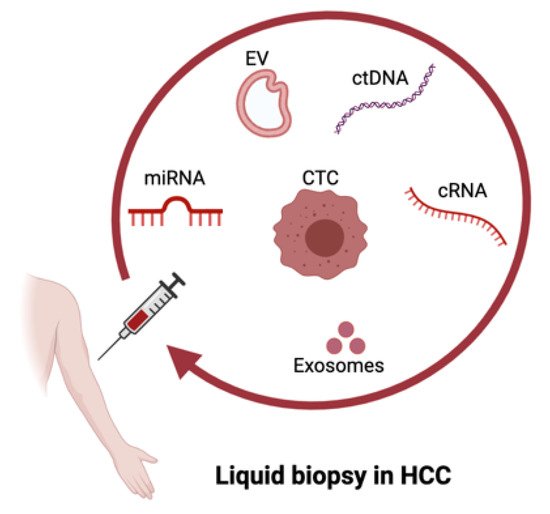
2.1. ctDNA in HCC
2.2. CTCs in HCC
CTCs are also emerging as a promising biomarker for the prediction of HCC treatment efficacies. CTCs arise in the circulation after detachment from primary or metastatic tumor lesions [44]. They differ from other types of cancer biomarkers as they represent viable tumor cells circulating in the patient’s bloodstream. Therefore, CTCs can also provide comprehensive genetic information about tumor heterogeneity and drug sensitivity [20]. CTCs have been approved by the FDA as diagnostic markers for specific epithelial cancers [16]. However, their diagnostic role in HCC still requires further studies. A widely known CTC biomarker is the epithelial cell adhesion molecule (EpCAM) [44], a pan-cancer biomarker which has also been observed in HCC patients [45].
Despite the highly promising role of CTCs as a biomarker for HCC [20], it remains challenging to detect HCC CTCs early and accurately because of the lack of specific markers. Another limitation is that the frequencies of CTCs are usually low in the circulation, especially at the early stages [16][20]. Thus, combinational strategies may be needed, in order to improve the prognostic and diagnostic value of HCC.
3. Liquid Biopsy as a Diagnostic and Prognostic Tool
Liquid biopsy has been explored as a way to monitor cancer prognosis and diagnosis in a non-invasive manner. This technology has shown promising results in early diagnosis [46], detection of minimal residual disease [47] and decision making for systemic therapies of different types of cancers, including HCC [8][48][49].
Among all liquid biopsy analytes, ctDNA plays an important role in HCC prognosis [17]. ctDNA maintains the same genomic signatures that are present in the matching tumor tissue, allowing for the quantitative and qualitative evaluation of the mutation burden in body fluids [50]. In this way, ctDNA has been considered as a good biomarker and can be utilized in disease monitoring. The data of ctDNA include quantitative changes, such as differences in the concentration of ctDNA, as well as qualitative changes, such as gene mutations, DNA copy number variations and DNA methylation [16]. Indicatively, studies based on the detection of somatic single-nucleotide mutations and methylation changes in ctDNA could closely correlate with tumor burden over time in HCC patients and could be used to predict recurrence after surgery [17][51][52].
As ctDNA represents only a very small proportion of cell-free DNA, very sensitive and reliable detection methods are required. Levels of ctDNA are measured mainly by real-time PCR (RT-PCR) [53], while digital PCR (dPCR) [54] or sequencing methods are used for the detection of point mutations [55]. In addition to TERT and TP53 mutations as the prognostic factors of poor survival [56][57], other mutations have been shown to have prognostic values for HCC. MLH1 mutation was specifically associated with lower survival [1], whereas mutations of genes from the PI3K/mTOR pathway were shown to be the predictors of non-responders to TKI treatments for patients with advanced HCC [49].
A number of studies have also shown the prognostic values of circulating miRNAs in HCC. Lower survival rates were associated with patients with low levels of miR-1, miR-122, miR-26a, miR-29a and miR-223-3p [58][59][60][61] or high levels of miR-155, miR-96 and miR-193-5p [62][63]. Furthermore, six additional miRNAs were identified as prognostic factors. Low levels of miR-424-5p or miR- 101-3p and high levels of miR-128, miR-139-5p, miR-382-5p and miR410 were associated with lower survival rates in HCC patients [17]. Alternatively, miRNAs have been studied in association with EVs [32][33][34]. In a cohort of 59 HCC patients, a correlation was found between tumor recurrences after liver transplantation and a high level of exosomal miR-718 [64]. Additionally, high levels of exosomal miR-665 or low levels of exosomal miR-638 and miR-320a were identified as predictors of poor survival [65][66][67].
Another cornerstone of liquid biopsy is the isolation and detection of CTCs, which have been described as a useful tool for the prognostication of HCC [68]. As introduced above, EpCAM-positive CTC cells have been intensively investigated in HCC [45][69]. However, since CTCs can lose their epithelial phenotype through epithelial-to-mesenchymal transition (EMT) in order to survive and metastasize [44], EpCAM cannot always be considered an optimal biomarker to detect HCC. Alternatively, other phenotypic markers have been explored, such as the hepatocyte-specific asialoglycoprotein receptor (ASGPR) [70], and the hepatocyte paraffin 1 [71], or incorporation of several markers simultaneously, as it has extensively been reviewed elsewhere [20]. Most recently, in a prospective study of 80 HCC patients, a multimarker assay combining cell surface markers EpCAM, ASGPR and GPC3 was able to detect HCC CTCs in 97% of the patients with high accuracy. Moreover, a phenotypic variant subset of CTCs was associated with aggressive disease progression and underlying metastasis, therefore highlighting the important implications of CTCs in treatment selection [72]. Another study showed that the detection of phosphorylated ERK (pERK) and pAkt in CTCs could predict the response to sorafenib efficacy in advanced HCC patients, similarly to tumor tissue biopsy [73].
4. Liquid Biopsy for Immunotherapy in HCC
The race towards the identification of immunotherapy predictive biomarkers is at the forefront of research in HCC. Among the biomarkers of interest, there are TMB and mutational signatures identified from ctDNA, and PD-L1 expression detected on CTCs [74]. TMB and PD-L1 expression are considered good predictors in several cancers, but the evidence in liver cancer has not been as established thus far [75]. In Table 1, we summarize the most recent literature in the field, and we highlight the key findings for each study.
| Type of Biomarker Analyzed | Key Findings | Reference |
|---|---|---|
| Changes in the ctDNA levels | Could significantly correlate with tumor size in cancer patients treated with anti-PD1 drugs and be a valuable prognostic factor of progression-free and overall survival. | [76] |
| Targeted gene analysis of ctDNA | Can be a better option to evaluate TMB prior to immunotherapy in cases of advanced primary liver cancers when tissue biopsy is not recommended. | [18] |
| Mutational analysis of ctDNA | Could not be associated with response to ICI therapy but only to systemic treatment. | [36] |
| Levels of ctDNA at baseline | Higher levels of ctDNA at baseline were associated with an increased baseline tumor burden, and ctDNA turned negative in 70%, 27%, 9% and 0% of patients achieving a complete response, partial response, stable disease and disease progression, respectively. | [77] |
| Undetectable ctDNA levels during treatment were linked to a longer progression-free survival. | ||
| Hyper-mutated ctDNA phenotype | Is associated with a favorable outcome in a cohort of 69 cancer patients with different histologies, including three HCC patients, treated with different immune checkpoint inhibitors. | [78] |
| Overall response rate, PFS and OS in high-alteration groups were significantly higher than in low-alteration groups. | ||
| Detection of Wnt/b-catenin-activating mutations | Wnt/b-catenin-activating mutations in HCC linked to potential tumor immunotherapy resistance in several studies. | [79][80][81][82] |
| Detection of Wnt/b-catenin-activating mutations | Demonstration that liquid biopsy is potentially able to detect Wnt/b-catenin-activating mutations in HCC. | [75] |
| Detection of Wnt/b-catenin-activating mutations | Detection of Wnt/b-catenin pathway-activating mutations might not be sufficient to identify advanced HCC patients with primary resistance to ICI. | [83] |
| Targeted mutational analysis of CTNNB1 p.T41A mutation | ctDNA liquid biopsy managed to reveal mutations that were not detected in single tumor biopsies, thus increasing the detection rate of CTNNB1 mutation in HCC patients. | [84] |
| PD-L1 expression on CTCs | Biomarker to assess ICI-based immunotherapy efficacy of advanced solid tumors. | [85] |
| CTCs expressing PD-L1 | PD-L1-positive CTCs are mainly found in advanced stages of disease, and they represent an independent prognostic factor for overall survival. 6 out of 10 patients receiving anti-PD-1 treatment had PD-L1-positive CTCs at baseline, and of these, 5 out of 6 had a favorable treatment response. 4 out of 10 patients receiving anti-PD-1 treatment did not have PD-L1+ CTCs and were non-responders. |
[86] |
5. Limitations and Future Perspectives
References
- Kim, E.; Viatour, P. Hepatocellular carcinoma: Old friends and new tricks. Exp. Mol. Med. 2020, 52, 1898–1907.
- Lai, E.; Astara, G.; Ziranu, P.; Pretta, A.; Migliari, M.; Dubois, M.; Donisi, C.; Mariani, S.; Liscia, N.; Impera, V.; et al. Introducing immunotherapy for advanced hepatocellular carcinoma patients: Too early or too fast? Crit. Rev. Oncol. Hematol. 2021, 157, 103167.
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314.
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.-Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905.
- Sangro, B.; Sarobe, P.; Hervás-Stubbs, S.; Melero, I. Advances in immunotherapy for hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 525–543.
- El-Khoueiry, A.B.; Sangro, B.; Yau, T.; Crocenzi, T.S.; Kudo, M.; Hsu, C.; Kim, T.-Y.; Choo, S.-P.; Trojan, J.; Welling, T.H.; et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): An open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 2017, 389, 2492–2502.
- Davis, A.A.; Patel, V.G. The role of PD-L1 expression as a predictive biomarker: An analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J. Immunother. Cancer 2019, 7, 1–8.
- Lee, E.Y.; Kulkarni, R.P. Circulating biomarkers predictive of tumor response to cancer immunotherapy. Expert Rev. Mol. Diagn. 2019, 19, 895–904.
- Goodman, A.M.; Kato, S.; Bazhenova, L.; Patel, S.P.; Frampton, G.M.; Miller, V.; Stephens, P.J.; Daniels, G.A.; Kurzrock, R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol. Cancer Ther. 2017, 16, 2598–2608.
- Kowanetz, M.; Zou, W.; Gettinger, S.N.; Koeppen, H.; Kockx, M.; Schmid, P.; Kadel, E.E.; Wistuba, I., 3rd; Chaft, J.; Rizvi, N.A.; et al. Differential regulation of PD-L1 expression by immune and tumor cells in NSCLC and the response to treatment with atezolizumab (anti-PD-L1). Proc. Natl. Acad. Sci. USA 2018, 115, e10119–e10126.
- Herbst, R.S.; Baas, P.; Kim, D.-W.; Felip, E.; Perez-Gracia, J.L.; Han, J.-Y.; Molina, J.; Kim, J.-H.; Arvis, C.D.; Ahn, M.-J.; et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet 2015, 387, 1540–1550.
- Hoshida, Y.; Villanueva, A.; Kobayashi, M.; Peix, J.; Chiang, D.; Camargo, A.; Gupta, S.; Moore, J.; Wrobel, M.J.; Lerner, J.; et al. Gene Expression in Fixed Tissues and Outcome in Hepatocellular Carcinoma. N. Engl. J. Med. 2008, 359, 1995–2004.
- Quandt, D.; Zucht, H.D.; Amann, A.; Wulf-Goldenberg, A.; Borrebaeck, C.; Cannarile, M.; Lambrechts, D.; Oberacher, H.; Garrett, J.; Nayak, T.; et al. Implementing liquid biopsies into clinical decision making for cancer immunotherapy. Oncotarget 2017, 8, 48507–48520.
- Cheng, J.; Li, M.; Lv, Y. Sublethal heat treatment promotes epithelial-mesenchymal transition and enhances the malignant potential of hepatocellular carcinoma. Hepatology 2013, 59, 1650.
- Jamal-Hanjani, M.; Quezada, S.; Larkin, J.; Swanton, C. Translational Implications of Tumor Heterogeneity. Clin. Cancer Res. 2015, 21, 1258–1266.
- Mocan, T.; Simao, A.L.; Castro, R.E.; Rodrigues, C.M.P.; Slomka, A.; Wang, B.; Strassburg, C.; Wohler, A.; Willms, A.G.; Kornek, M. Liquid Biopsies in Hepatocellular Carcinoma: Are We Winning? J. Clin. Med. 2020, 9, 1541.
- Labgaa, I.; Villanueva, A.; Dormond, O.; Demartines, N.; Melloul, E. The Role of Liquid Biopsy in Hepatocellular Carcinoma Prognostication. Cancers 2021, 13, 659.
- Zhao, W.; Qiu, L.; Liu, H.; Xu, Y.; Zhan, M.; Zhang, W.; Xin, Y.; He, X.; Yang, X.; Bai, J.; et al. Circulating tumor DNA as a potential prognostic and predictive biomarker during interventional therapy of unresectable primary liver cancer. J. Gastrointest. Oncol. 2020, 11, 1065–1077.
- Silva, M.A.; Hegab, B.; Hyde, C.; Guo, B.; Buckels, J.A.C.; Mirza, D.F. Needle track seeding following biopsy of liver lesions in the diagnosis of hepatocellular cancer: A systematic review and meta-analysis. Gut 2008, 57, 1592–1596.
- Chen, F.; Zhong, Z.; Tan, H.-Y.; Wang, N.; Feng, Y. The Significance of Circulating Tumor Cells in Patients with Hepatocellular Carcinoma: Real-Time Monitoring and Moving Targets for Cancer Therapy. Cancers 2020, 12, 1734.
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930.
- Li, J.; Han, X.; Yu, X.; Xu, Z.; Yang, G.; Liu, B.; Xiu, P. Clinical applications of liquid biopsy as prognostic and predictive biomarkers in hepatocellular carcinoma: Circulating tumor cells and circulating tumor DNA. J. Exp. Clin. Cancer Res. 2018, 37, 1–13.
- Ryder, S.D. Guidelines for the diagnosis and treatment of hepatocellular carcinoma (HCC) in adults. Gut 2003, 52, 1–8.
- Alunni-Fabbroni, M.; Rönsch, K.; Huber, T.; Cyran, C.C.; Seidensticker, M.; Mayerle, J.; Pech, M.; Basu, B.; Verslype, C.; Benckert, J.; et al. Circulating DNA as prognostic biomarker in patients with advanced hepatocellular carcinoma: A translational exploratory study from the SORAMIC trial. J. Transl. Med. 2019, 17, 1–15.
- Huang, A.; Zhang, X.; Zhou, S.-L.; Cao, Y.; Huang, X.-W.; Fan, J.; Yang, X.-R.; Zhou, J. Plasma Circulating Cell-free DNA Integrity as a Promising Biomarker for Diagnosis and Surveillance in Patients with Hepatocellular Carcinoma. J. Cancer 2016, 7, 1798–1803.
- Xiong, Y.; Xie, C.-R.; Zhang, S.; Chen, J.; Yin, Z.-Y. Detection of a novel panel of somatic mutations in plasma cell-free DNA and its diagnostic value in hepatocellular carcinoma. Cancer Manag. Res. 2019, 11, 5745–5756.
- El-Tawdi, A.H.F.; Matboli, M.; Shehata, H.H.; Tash, F.; El-Khazragy, N.; Azazy, A.E.-S.M.; Abdel-Rahman, O. Evaluation of Circulatory RNA-Based Biomarker Panel in Hepatocellular Carcinoma. Mol. Diagn. Ther. 2016, 20, 265–277.
- Moshiri, F.; Salvi, A.; Gramantieri, L.; Sangiovanni, A.; Guerriero, P.; De Petro, G.; Bassi, C.; Lupini, L.; Sattari, A.; Cheung, D.; et al. Circulating miR-106b-3p, miR-101-3p and miR-1246 as diagnostic biomarkers of hepatocellular carcinoma. Oncotarget 2018, 9, 15350–15364.
- Yamamoto, Y.; Kondo, S.; Matsuzaki, J.; Esaki, M.; Okusaka, T.; Shimada, K.; Murakami, Y.; Enomoto, M.; Tamori, A.; Kato, K.; et al. Highly Sensitive Circulating MicroRNA Panel for Accurate Detection of Hepatocellular Carcinoma in Patients With Liver Disease. Hepatol. Commun. 2019, 4, 284–297.
- Kelley, R.K.; Magbanua, M.J.M.; Butler, T.M.; Collisson, E.A.; Hwang, J.; Sidiropoulos, N.; Evason, K.; McWhirter, R.M.; Hameed, B.; Wayne, E.M.; et al. Circulating tumor cells in hepatocellular carcinoma: A pilot study of detection, enumeration, and next-generation sequencing in cases and controls. BMC Cancer 2015, 15, 1–11.
- Von Felden, J.; Schulze, K.; Krech, T.; Ewald, F.; Nashan, B.; Pantel, K.; Lohse, A.W.; Riethdorf, S.; Wege, H. Circulating tumor cells as liquid biomarker for high HCC recurrence risk after curative liver resection. Oncotarget 2017, 8, 89978–89987.
- Julich-Haertel, H.; Urban, S.K.; Krawczyk, M.; Willms, A.; Jankowski, K.; Patkowski, W.; Kruk, B.; Krasnodębski, M.; Ligocka, J.; Schwab, R.; et al. Cancer-associated circulating large extracellular vesicles in cholangiocarcinoma and hepatocellular carcinoma. J. Hepatol. 2017, 67, 282–292.
- Wang, Y.; Zhang, C.; Zhang, P.; Guo, G.; Jiang, T.; Zhao, X.; Jiang, J.; Huang, X.; Tong, H.; Tian, Y. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med. 2018, 7, 1670–1679.
- Tucci, M.; Passarelli, A.; Mannavola, F.; Stucci, L.S.; Ascierto, P.A.; Capone, M.; Madonna, G.; Lopalco, P.; Silvestris, F. Serum exosomes as predictors of clinical response to ipilimumab in metastatic melanoma. OncoImmunology 2017, 7, e1387706.
- Chakrabarti, S.; Xie, H.; Urrutia, R.; Mahipal, A. The Promise of Circulating Tumor DNA (ctDNA) in the Management of Early-Stage Colon Cancer: A Critical Review. Cancers 2020, 12, 2808.
- von Felden, J.; Craig, A.J.; Garcia-Lezana, T.; Labgaa, I.; Haber, P.K.; D’Avola, D.; Asgharpour, A.; Dieterich, D.; Bonaccorso, A.; Torres-Martin, M.; et al. Mutations in circulating tumor DNA predict primary resistance to systemic therapies in advanced hepatocellular carcinoma. Oncogene 2020, 40, 140–151.
- Cai, J.; Chen, L.; Zhang, Z.; Zhang, X.; Lu, X.; Liu, W.; Shi, G.; Ge, Y.; Gao, P.; Yang, Y.; et al. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut 2019, 68, 2195–2205.
- Xu, R.-H.; Wei, W.; Krawczyk, M.; Wang, W.; Luo, H.; Flagg, K.; Yi, S.; Shi, W.; Quan, Q.; Li, K.; et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 2017, 16, 1155–1161.
- Kaseb, A.O.; Sánchez, N.S.; Sen, S.; Kelley, R.K.; Tan, B.R.; Bocobo, A.G.; Lim, K.H.; Abdel-Wahab, R.; Uemura, M.; Pestana, R.C.; et al. Molecular Profiling of Hepatocellular Carcinoma Using Circulating Cell-Free DNA. Clin. Cancer Res. 2019, 25, 6107–6118.
- Liao, W.; Yang, H.; Xu, H.; Wang, Y.; Ge, P.; Ren, J.; Xu, W.; Lü, X.; Sang, X.; Zhong, S.; et al. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepatocellular carcinoma patients by targeted deep sequencing. Oncotarget 2016, 7, 40481–40490.
- Chan, K.C.A.; Lai, P.B.-S.; Mok, T.; Chan, H.L.Y.; Ding, C.; Yeung, S.W.; Lo, Y.M.D. Quantitative Analysis of Circulating Methylated DNA as a Biomarker for Hepatocellular Carcinoma. Clin. Chem. 2008, 54, 1528–1536.
- Tran, N.H.; Kisiel, J.; Roberts, L.R. Using cell-free DNA for HCC surveillance and prognosis. JHEP Rep. 2021, 3.
- David, W.; Cescon, S.V.B.; Steven, M. Chan and Lillian, L. Siu1 Circulating tumor DNA and liquid biopsy in oncology. Nat. Cncer 2020, 1, 276–290.
- Plaks, V.; Koopman, C.D.; Werb, Z. Cancer. Circulating tumor cells. Science 2013, 341, 1186–1188.
- Guo, W.; Yang, X.R.; Sun, Y.F.; Shen, M.N.; Ma, X.L.; Wu, J.; Zhang, C.Y.; Zhou, Y.; Xu, Y.; Hu, B.; et al. Clinical significance of EpCAM mRNA-positive circulating tumor cells in hepatocellular carcinoma by an optimized negative enrichment and qRT-PCR-based platform. Clin. Cancer Res. 2014, 20, 4794–4805.
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224ra24.
- Tie, J.; Wang, Y.; Tomasetti, C.; Li, L.; Springer, S.; Kinde, I.; Silliman, N.; Tacey, M.; Wong, H.-L.; Christie, M.; et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016, 8, 346ra92.
- Labgaa, I.; Villacorta-Martin, C.; D’Avola, D.; Craig, A.J.; Von Felden, J.; Filho, S.M.; Sia, D.; Stueck, A.; Ward, S.C.; Fiel, M.I.; et al. A pilot study of ultra-deep targeted sequencing of plasma DNA identifies driver mutations in hepatocellular carcinoma. Oncogene 2018, 37, 3740–3752.
- Von Felden, J.; Garcia-Lezana, T.; Schulze, K.; Losic, B.; Villanueva, A. Liquid biopsy in the clinical management of hepatocellular carcinoma. Gut 2020, 69, 2025–2034.
- Vymetalkova, V.; Cervena, K.; Bartu, L.; Vodicka, P. Circulating Cell-Free DNA and Colorectal Cancer: A Systematic Review. Int. J. Mol. Sci. 2018, 19, 3356.
- Ono, A.; Fujimoto, A.; Yamamoto, Y.; Akamatsu, S.; Hiraga, N.; Imamura, M.; Kawaoka, T.; Tsuge, M.; Abe, H.; Hayes, C.N.; et al. Circulating Tumor DNA Analysis for Liver Cancers and Its Usefulness as a Liquid Biopsy. Cell. Mol. Gastroenterol. Hepatol. 2015, 1, 516–534.
- Cai, Z.; Chen, G.; Zeng, Y.; Dong, X.; Li, Z.; Huang, Y.; Xin, F.; Qiu, L.; Xu, H.; Zhang, W.; et al. Comprehensive Liquid Profiling of Circulating Tumor DNA and Protein Biomarkers in Long-Term Follow-Up Patients with Hepatocellular Carcinoma. Clin. Cancer Res. 2019, 25, 5284–5294.
- Iizuka, N.; Sakaida, I.; Moribe, T.; Fujita, N.; Miura, T.; Stark, M.; Tamatsukuri, S.; Ishitsuka, H.; Uchida, K.; Terai, S.; et al. Elevated levels of circulating cell-free DNA in the blood of patients with hepatitis C virus-associated hepatocellular carcinoma. Anticancer. Res. 2007, 26, 4713–4719.
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610.
- Chan, K.C.A.; Jiang, P.; Chan, C.W.M.; Sun, K.; Wong, J.; Hui, E.P.; Chan, S.; Chan, W.C.; Hui, D.; Ng, S.S.M.; et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 18761–18768.
- Oversoe, S.K.; Clement, M.S.; Pedersen, M.H.; Weber, B.; Aagaard, N.K.; Villadsen, G.E.; Grønbæk, H.; Hamilton-Dutoit, S.J.; Sorensen, B.S.; Kelsen, J. TERT promoter mutated circulating tumor DNA as a biomarker for prognosis in hepatocellular carcinoma. Scand. J. Gastroenterol. 2020, 55, 1433–1440.
- Shen, T.; Li, S.; Wang, J.; Zhang, T.; Zhang, S.; Chen, H.; Xiao, Q.; Ren, W.; Liu, C.; Peng, B.; et al. TP53 R249S mutation detected in circulating tumour DNA is associated with Prognosis of hepatocellular carcinoma patients with or without hepatectomy. Liver Int. 2020, 40, 2834–2847.
- Köberle, V.; Kronenberger, B.; Pleli, T.; Trojan, J.; Imelmann, E.; Peveling-Oberhag, J.; Welker, M.-W.; Elhendawy, M.; Zeuzem, S.; Piiper, A.; et al. Serum microRNA-1 and microRNA-122 are prognostic markers in patients with hepatocellular carcinoma. Eur. J. Cancer 2013, 49, 3442–3449.
- Xu, Y.; Bu, X.; Dai, C.; Shang, C. High serum microRNA-122 level is independently associated with higher overall survival rate in hepatocellular carcinoma patients. Tumor Biol. 2015, 36, 4773–4776.
- Cho, H.J.; Kim, S.S.; Nam, J.S.; Kim, J.K.; Lee, J.H.; Kim, B.; Wang, H.J.; Kim, B.W.; Lee, J.D.; Kang, D.Y.; et al. Low levels of circulating microRNA-26a/29a as poor prognostic markers in patients with hepatocellular carcinoma who underwent curative treatment. Clin. Res. Hepatol. Gastroenterol. 2017, 41, 181–189.
- Pratedrat, P.; Chuaypen, N.; Nimsamer, P.; Payungporn, S.; Pinjaroen, N.; Sirichindakul, B.; Tangkijvanich, P. Diagnostic and prognostic roles of circulating miRNA-223-3p in hepatitis B virus–related hepatocellular carcinoma. PLoS ONE 2020, 15, e0232211.
- Ning, S.; Liu, H.; Gao, B.; Wei, W.; Yang, A.; Li, J.; Zhang, J. miR-155, miR-96 and miR-99a as potential diagnostic and prognostic tools for the clinical management of hepatocellular carcinoma. Oncol. Lett. 2019, 18, 3381–3387.
- Loosen, S.H.; Wirtz, T.H.; Roy, S.; Vucur, M.; Castoldi, M.; Schneider, A.T.; Koppe, C.; Ulmer, T.F.; Roeth, A.A.; Bednarsch, J.; et al. Circulating levels of microRNA193a-5p predict outcome in early stage hepatocellular carcinoma. PLoS ONE 2020, 15, e0239386.
- Sugimachi, K.; Matsumura, T.; Hirata, H.; Uchi, R.; Ueda, M.; Ueo, H.; Shinden, Y.; Iguchi, T.; Eguchi, H.; Shirabe, K.; et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br. J. Cancer 2015, 112, 532–538.
- Qu, Z.; Wu, J.; Wu, J.; Ji, A.; Qiang, G.; Jiang, Y.; Jiang, C.; Ding, Y. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget 2017, 8, 80666–80678.
- Shi, M.; Jiang, Y.; Yang, L.; Yan, S.; Wang, Y.; Lu, X. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J. Cell. Biochem. 2017, 119, 4711–4716.
- Hao, X.; Xin, R.; Dong, W. Decreased serum exosomal miR-320a expression is an unfavorable prognostic factor in patients with hepatocellular carcinoma. J. Int. Med. Res. 2020, 48.
- Ye, Q.; Ling, S.; Zheng, S.; Xu, X. Liquid biopsy in hepatocellular carcinoma: Circulating tumor cells and circulating tumor DNA. Mol. Cancer 2019, 18, 1–13.
- Ahn, J.C.; Teng, P.-C.; Chen, P.; Posadas, E.; Tseng, H.; Lu, S.C.; Yang, J.D. Detection of Circulating Tumor Cells and Their Implications as a Biomarker for Diagnosis, Prognostication, and Therapeutic Monitoring in Hepatocellular Carcinoma. Hepatology 2020, 73, 422–436.
- Li, J.; Chen, L.; Zhang, X.; Zhang, Y.; Liu, H.; Sun, B.; Zhao, L.; Ge, N.; Qian, H.; Yang, Y.; et al. Detection of Circulating Tumor Cells in Hepatocellular Carcinoma Using Antibodies against Asialoglycoprotein Receptor, Carbamoyl Phosphate Synthetase 1 and Pan-Cytokeratin. PLoS ONE 2014, 9, e96185.
- Lamps, L.W.; Folpe, A.L. The Diagnostic Value of Hepatocyte Paraffin Antibody 1 in Differentiating Hepatocellular Neoplasms from Nonhepatic Tumors: A Review. Adv. Anat. Pathol. 2003, 10, 39–43.
- Court, C.M.; Hou, S.; Winograd, P.; Segel, N.H.; Li, Q.W.; Zhu, Y.; Sadeghi, S.; Finn, R.S.; Ganapathy, E.; Song, M.; et al. A novel multimarker assay for the phenotypic profiling of circulating tumor cells in hepatocellular carcinoma. Liver Transplant. 2018, 24, 946–960.
- Li, J.; Shi, L.; Zhang, X.; Sun, B.; Yang, Y.; Ge, N.; Liu, H.; Yang, X.; Chen, L.; Qian, H.; et al. pERK/pAkt phenotyping in circulating tumor cells as a biomarker for sorafenib efficacy in patients with advanced hepatocellular carcinoma. Oncotarget 2015, 7, 2646–2659.
- Armstrong, P.P.S.; Aiwu, R.H. Immunotherapy and immunotherapy biomarkers for hepatocellular carcinoma. Hepatoma Res. 2021, 7, 18.
- Kwee, S.A.; Tiirikainen, M. Beta-catenin activation and immunotherapy resistance in hepatocellular carcinoma: Mechanisms and biomarkers. Hepatoma Res. 2021, 2021.
- Cabel, L.; Riva, F.; Servois, V.; Livartowski, A.; Daniel, C.; Rampanou, A.; Lantz, O.; Romano, E.; Milder, M.; Buecher, B.; et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: A proof-of-concept study. Ann. Oncol. 2017, 28, 1996–2001.
- Hsu, C.-H.; Lu, S.; Abbas, A.; Guan, Y.; Zhu, A.X.; Aleshin, A.; Lee, K.-H.; Lee, M.S.; Mahipal, A.; Ryoo, B.-Y.; et al. Longitudinal and personalized detection of circulating tumor DNA (ctDNA) for monitoring efficacy of atezolizumab plus bevacizumab in patients with unresectable hepatocellular carcinoma (HCC). J. Clin. Oncol. 2020, 38, 3531.
- Khagi, Y.; Kurzrock, R.; Patel, S.P. Next generation predictive biomarkers for immune checkpoint inhibition. Cancer Metastas. Rev. 2016, 36, 179–190.
- Luke, J.J.; Bao, R.; Sweis, R.F.; Spranger, S.; Gajewski, T.F. WNT/β-catenin Pathway Activation Correlates with Immune Exclusion across Human Cancers. Clin. Cancer Res. 2019, 25, 3074–3083.
- Sia, D.; Jiao, Y.; Martinez-Quetglas, I.; Kuchuk, O.; Villacorta-Martin, C.; de Moura, M.C.; Putra, J.; Campreciós, G.; Bassaganyas, L.; Akers, N.; et al. Identification of an Immune-specific Class of Hepatocellular Carcinoma, Based on Molecular Features. Gastroenterology 2017, 153, 812–826.
- Harding, J.J.; Nandakumar, S.; Armenia, J.; Khalil, D.N.; Albano, M.; Ly, M.; Shia, J.; Hechtman, J.; Kundra, R.; El Dika, I.; et al. Prospective Genotyping of Hepatocellular Carcinoma: Clinical Implications of Next-Generation Sequencing for Matching Patients to Targeted and Immune Therapies. Clin. Cancer Res. 2018, 25, 2116–2126.
- De Galarreta, M.R.; Bresnahan, E.; Molina-Sánchez, P.; Lindblad, K.E.; Maier, B.; Sia, D.; Puigvehi, M.; Miguela, V.; Casanova-Acebes, M.; Dhainaut, M.; et al. β-Catenin Activation Promotes Immune Escape and Resistance to Anti–PD-1 Therapy in Hepatocellular Carcinoma. Cancer Discov. 2019, 9, 1124–1141.
- von Felden, J.; Villanueva, A. Role of Molecular Biomarkers in Liver Transplantation for Hepatocellular Carcinoma. Liver Transpl. 2020, 26, 823–831.
- Oversoe, S.K.; Clement, M.S.; Weber, B.; Grønbæk, H.; Hamilton-Dutoit, S.J.; Sorensen, B.S.; Kelsen, J. Combining tissue and circulating tumor DNA increases the detection rate of a CTNNB1 mutation in hepatocellular carcinoma. BMC Cancer 2021, 21, 1–7.
- Yue, C.; Jiang, Y.; Li, P.; Wang, Y.; Xue, J.; Li, N.; Li, D.; Wang, R.; Dang, Y.; Hu, Z.; et al. Dynamic change of PD-L1 expression on circulating tumor cells in advanced solid tumor patients undergoing PD-1 blockade therapy. OncoImmunology 2018, 7, e1438111.
- Winograd, P.; Hou, S.; Court, C.M.; Lee, Y.; Chen, P.; Zhu, Y.; Sadeghi, S.; Finn, R.S.; Teng, P.; Wang, J.J.; et al. Hepatocellular Carcinoma–Circulating Tumor Cells Expressing PD-L1 Are Prognostic and Potentially Associated with Response to Checkpoint Inhibitors. Hepatol. Commun. 2020, 4, 1527–1540.
- Chen, V.L.; Xu, D.; Wicha, M.S.; Lok, A.S.; Parikh, N.D. Utility of Liquid Biopsy Analysis in Detection of Hepatocellular Carcinoma, Determination of Prognosis, and Disease Monitoring: A Systematic Review. Clin. Gastroenterol. Hepatol. 2020, 18, 2879–2902.e9.
- McDonald, B.R.; Contente-Cuomo, T.; Sammut, S.-J.; Odenheimer-Bergman, A.; Ernst, B.; Perdigones, N.; Chin, S.-F.; Farooq, M.; Mejia, R.; Cronin, P.A.; et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci. Transl. Med. 2019, 11, eaax7392.
- Dhar, M.; Wong, J.; Che, J.; Matsumoto, M.; Grogan, T.; Elashoff, D.; Garon, E.B.; Goldman, J.W.; Christen, E.S.; Di Carlo, D.; et al. Evaluation of PD-L1 expression on vortex-isolated circulating tumor cells in metastatic lung cancer. Sci. Rep. 2018, 8, 1–10.
- Krieg, C.; Nowicka, M.; Guglietta, S.; Schindler, S.; Hartmann, F.J.; Weber, L.M.; Dummer, R.; Robinson, M.D.; Levesque, M.P.; Becher, B. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat. Med. 2018, 24, 144–153.
- Masucci, G.V.; Cesano, A.; Hawtin, R.; Janetzki, S.; Zhang, J.; Kirsch, I.; Dobbin, K.K.; Alvarez, J.; Robbins, P.B.; Selvan, S.R.; et al. Validation of biomarkers to predict response to immunotherapy in cancer: Volume I—Pre-analytical and analytical validation. J. Immunother. Cancer 2016, 4, 76.
- Merle, P. The New Immuno-Oncology-Based Therapies and Their Perspectives in Hepatocellular Carcinoma. Cancers 2021, 13, 238.




