2.1. Apoptotic Engulfment in Mammals
The efficient clearance of senescent and dying cells during ontogenesis and aging is crucial for the maintenance of tissue homeostasis. The phagocytosis of apoptotic cells, termed efferocytosis, is considered to be immunologically silent or even immune-suppressive, with tissue-resident macrophages releasing anti-inflammatory mediators and dendritic cells inducing the differentiation of tolerogenic T cells after the uptake of apoptotic cells
[3][4][5].
Although cells undergoing apoptosis retain an intact plasma membrane, there are changes in the phospholipid composition in the outer leaflet that work as an “eat me” signal for phagocytes. The most well-known indicator of apoptosis is the exposure of phosphatidylserine (PtdSer) on the cell surface
[6]. In healthy cells, this phospholipid is actively kept in the inner leaflet by lipid transporters called flippases
[7]. The caspases present during apoptosis both disrupt the function of flippases maintaining the asymmetrical distribution of PtdSer and activate scramblase enzymes that catalyse the reverse translocation of this molecule
[8][9].
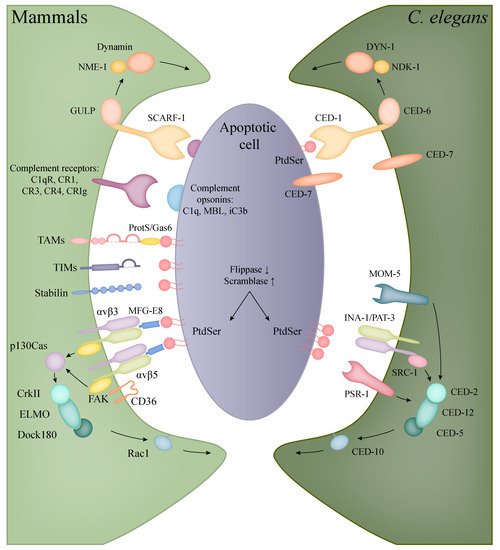
PtdSer is either directly recognised or is bound to receptors on phagocytes through soluble bridging molecules (
Figure 2,
Table 1). A PtdSer receptor (PSR) was first described by Fadok et al. using a monoclonal antibody (mAb217) that bound to the surface of macrophages and inhibited the uptake of apoptotic bodies
[10]. However, subsequent experiments trying to identify this receptor produced some contradictory data
[11]. The protein encoded by the
psr gene, later named JMJD6 (Jumonji domain-containing protein 6), was shown to be located in the nucleus, having an important function in embryonic development with a histone demethylase activity
[12][13]. Thus, it is possible that the surface molecule responsible for the binding of apoptotic bodies is not identical to the product of the suspected evolutionarily conserved
psr-1 gene.
Figure 2. Recognition and engulfment of apoptotic cells in mammalian phagocytes and C. elegans. The evolutionarily conserved CED-1/CED-6/DYN-1 (corresponding to human SCARF-1/GULP/Dynamin) and CED-2/CED-5/CED-12/CED-10 (in human: CrkII/Dock180/ELMO/Rac1) pathways are present in both C. elegans and mammals with the same functions in the clearance of apoptotic cells (orthologous molecules are indicated with the same colours). In mammals, however, a higher diversity can be observed in the recognition of apoptotic bodies, including the families of TIM and TAM proteins, complement and scavenger receptors.
Table 1. Molecules involved in the phagocytosis of apoptotic cells in human and C. elegans.
| Human Protein |
Function |
C. elegans Protein |
| ATP8A2 |
P4-type ATPase/flippase |
TAT-1 |
| XKR8 |
scramblase |
CED-8 |
| SCARF1, MEGF10 and 11, LRP1 (CD91), Jedi-1 |
phagocytic receptor |
CED-1 |
| GULP |
adaptor |
CED-6 |
| NME1 |
nucleoside diphosphate kinase |
NDK-1 |
| Dynamin |
large GTPase |
DYN-1 |
| ABCA1 and ABCA7 |
ABC transporter |
CED-7 |
| JMJD6 (PSR?) |
receptor |
PSR-1 |
| FZD1 and 7 (Frizzled class receptor 1 and 7) |
receptor |
MOM-5 |
| integrin α/β chain |
receptor |
INA-1/PAT-3 |
| SRC |
non-receptor tyrosine kinase |
SRC-1 |
| CrkII |
adaptor |
CED-2 |
| ELMO |
adaptor |
CED-12 |
| Dock180 |
Rac GEF |
CED-5 |
| Rac1 |
Rho family GTPase |
CED-10 |
| MFG-E8 |
bridging between PtdSer and integrins |
- |
| TIM1, 3, 4 |
PtdSer receptor |
- |
| Protein S, GAS6 |
bridging between PtdSer and TAM receptors (MER, AXL, TYRO3) |
- |
The receptors of the TIM family (T cell immunoglobulin- and mucin-domain-containing molecule) mediate the recognition and/or uptake of apoptotic bodies in various cell types by directly binding PtdSer. All three human TIM proteins (TIM1, -3, -4) have been proven to bind PtdSer, but the internalization of particles can be cell type-specific. TIM1 functions on T
h2-cells as a costimulatory molecule
[14] and it is upregulated in kidney epithelial cells after injury, allowing the engulfment of apoptotic bodies for tissue recovery
[15]. TIM3 is expressed on T
h1 cells and dendritic cells in humans, but the internalization of PtdSer containing apoptotic bodies was only observed in myeloid cells
[16][17][18]. Presumably, the most efficient apoptotic cell receptor of this family is TIM4 that is expressed on dendritic cells and various tissue-resident macrophages, like peritoneal or liver Kupffer cells, providing clearance throughout the body
[19][20][21].
The indirect binding of PtdSer can happen through the bridging molecules Protein S and GAS6 (growth-arrest-specific 6) and their receptors, the tyrosine kinase TAMs (Tyro3, Axl, Mer)
[22]. Protein S is synthetised mostly in liver hepatocytes and activates Tyro3 and Mer, but not Axl, whereas GAS6 acts as an agonist for all three TAM receptors
[23]. The TAM receptors are expressed in both macrophages and dendritic cells, but Seitz et al. showed that DCs mainly rely on the functions of Axl and Tyro3, whereas macrophages use all three
[24]. The signalling through TAMs contributes to tissue homeostasis via the inhibition of inflammatory responses in myeloid cells
[25][26].
Another group of soluble PtdSer-recognizing molecules includes MFG-E8 (milk fat globule-EGF factor 8 or lactadherin) and thrombospondin, which contain RGD motifs, thus connecting apoptotic cells to the integrins α
vβ
3 and α
vβ
5 [27][28]. Both integrins were shown to cooperate with the scavenger receptor CD36 in the uptake of apoptotic cells
[29][30]. The binding of apoptotic cells through these integrins initiates the phosphorylation of FAK and the recruitment of p130
Cas (Crk-associated SRC substrate), which connects to the evolutionary conserved CrkII–Dock180-ELMO-Rac1 pathway mediating the engulfment of the particles through actin polymerisation and phagocytic cup formation
[31][32][33]. Other scavenger receptors are also known to participate in the elimination of apoptotic cells, the specific ligands and functions of this receptor family are detailed in
Section 4.1 of the original article.
The complement system also participates in the opsonization, recognition and elimination of apoptotic cells. The complement components C1q and MBL bind apoptotic and necrotic cells, enhancing their uptake by phagocytes and in the presence of serum, activating the classical and lectin pathway
[34][35][36]. The activation of the alternative pathway by apoptotic cells was also proved by several studies, resulting in the deposition of C3 fragments and phagocytosis by the complement receptors CR1, CR3, CR4 and CRIg
[37][38][39]. Opsonization with complement facilitates the quick and effective clearance of apoptotic cells without an inflammatory response, which is further supported by the immunosuppressive qualities of C1q
[40][41].
2.2. Apoptotic Engulfment and Clearance in C. elegans
Programmed cell death can be divided into three phases: the first is the specification of the cell destined to die, and the second is the killing phase, where the apoptotic pathway is activated and lastly the execution of cell death and dismantling. The last phase also includes the elimination of apoptotic bodies, where a specialized or neighbouring cell engulfs and removes the debris of the dying cell. The conserved genes involved in the core apoptotic pathway were first identified in
C. elegans, for which the Nobel prize for Medicine was awarded to Sydney Brenner, John E. Sulston, and H. Robert Horvitz in 2002
[42][43].
Phagocytosis of apoptotic corpses involves conserved pathways in the nematode (
Table 2). The worm does not possess dedicated macrophages; instead, the neighbouring cells provide the phagocytic function. Similar to other organisms, the well-conserved PtdSer acts as the main signal of cell death or “eat-me” signal. The exposure of PtdSer in the worm during apoptosis is linked to the amino phospholipid translocase TAT-1 (Transbilayer Amphipath Transporter, ortholog of human flippase, ATPase phospholipid transporting 8A2, ATP8A2) and CED-8 (CEll Death abnormality 8, ortholog of human scramblase XKR8) function. In a living cell translocases actively transport PtdSer from the outer to the inner leaflet of the plasma membrane, but in an apoptotic cell, the inactivation of translocases and the activation of scramblases results in PtdSer accumulation on the cell surface
[44]. After the recognition of an apoptotic cell, the activated receptors stimulate the extension of the engulfing cell’s membrane and the rearrangement of the actin cytoskeleton. Consequently, the phagocytic cell develops pseudopods around the dying cell. Just as the pseudopods fuse, the newly formed phagosome separates from the plasma membrane
[45].
Two main, partly overlapping and conserved signalling pathways control the engulfment of apoptotic cells in
C. elegans [46]. One contains the phagocytic receptor CED-1 (human SCARF-1), the adaptor CED-6 (GULP), the ABC transporter CED-7 (ABCA) and the large GTPase DYN-1 (Dynamin2). The second path entails the CED-2 (CrkII), CED-5 (Dock180), CED-10 (Rac1), CED-12 (ELMO) proteins, the latter being the counterpart of the human Rac signalling. These two pathways partly converge at CED-10 involved in actin polymerisation, regulating the required cytoskeleton rearrangement for engulfment
[47][48].
The first pathway is triggered by the phagocytic transmembrane receptor CED-1, which only appears on the surface of the engulfing cell. After ligand recognition, the amount of CED-1 increases in the engulfing cell’s plasma membrane that is in contact with the neighbouring dead cell, initiating the formation of the phagocytic cup. The PtdSer driven activation of CED-1 triggers the subsequent members of the signalling cascade, resulting in the growth of pseudopods
[49]. CED-7 has a dual role to mediate the eat-me signal, it is expressed in both the engulfed and the engulfing cell: CED-7 assists in the exposure of PtdSer and also helps CED-1 to capture the PtdSer signal
[50]. Then the activated CED-1 signal is transmitted by CED-6, partly activating CED-10, but mostly triggers through DYN-1 that regulates the extension of the engulfing cell membrane
[47].
DYN-1 is a large GTPase that has multiple roles related to membrane trafficking. During engulfment, the phagocytic cup is transiently enriched in DYN-1 that provides the surplus of membrane necessary for pseudopod extension by helping the recruitment of early endosomes and their fusion with the cell membrane
[51]. Furthermore, it also mediates the last event in the engulfment process, namely the fission of phagosomes from the plasma membrane
[52]. For these functions, DYN-1 requires high amounts of GTP, which is provided by NDK-1 (Nucleoside Diphosphate Kinase-1, ortholog of human NME1)
[53]. It is important to note that both DYN-1 and NDK-1 are detected on the surface of early phagosomes, indicating that they also have roles in the early steps of phagosome maturation
[51][53][54].
The second pathway has an equally important role in the engulfing process: next to the membrane extension detailed above, the rearrangement of actin cytoskeleton is also crucial for the internalization of apoptotic cells. The components of this pathway regulate CED-10 activation
[55]. The pathway is triggered by the recognition of PtdSer by three different receptors: integrins, PSR-1 and MOM-5. The core of this pathway is the CED-2/CED-5/CED-12 trio, which complex acts as the guanine nucleotide exchange factor (GEF) of CED-10. The first possible route to trigger this pathway is through the integrin consisting of two subunits: INA-1 (integrin alpha-1) and PAT-3 (Paralysed Arrest at Two-fold, beta subunit)
[56][57]. In this case, CED-2 connects to the integrin receptor through SRC-1 (SRC oncogene related) and recruits the other two members of its complex
[56]. The other alpha subunit, PAT-2, might also have a role in recognising PtdSer, but the results published about the exact role of the PAT-2/PAT-3 heterodimer receptor in phagocytosis is controversial
[58][59]. The next receptor that has an influence on this pathway is PSR-1 (ortholog of human PSR, phosphatidyl serine receptor). CED-12 directly connects to this receptor and recruits the other two members of its complex upon activation
[57]. Besides the above-mentioned two receptors, the Frizzled homolog MOM-5 (More Of MS 5) connects the Wnt signalling to the engulfment process, which also acts through the CED-2/CED-5/CED-12 complex
[60].
Recently, a third pathway has been identified, revealing the important role of RAB-35 in the early steps of apoptotic cell phagocytosis. RAB-35 is a multifunctional GTPase that plays important roles in phagocytosis, cytokinesis, apico-basal polarity, cell migration, neurite outgrowth, immune synapse, exosome release and pathogen hijacking
[61]. During phagocytosis, RAB-35 localizes at the developing pseudopods, and later on early phagosomes, suggesting a function in early phagocytic events
[62]. The Zhou laboratory identified a novel role for RAB-35:
rab-35 mutants show a delay in apoptotic cell recognition, also
rab-35 mutant phenotypes are enhanced in both
ced-1 and
ced-5 mutant backgrounds, which is further worsened in
ced-1,
rab-35,
ced-5 triple mutants. As a conclusion,
rab-35 functions in a third pathway in parallel to the
ced-2/-5/-10/-12 and
ced-1/-6/-7 pathways, and further genetic epistasis analysis indicates that
rab-35 function is also linked to the integrin pathway
[62].
2.3. The Interconnection of Innate Immunity with Apoptotic Cell Clearance and the Nervous System
Phagocytes perform a dual role in host defence as they are both responsible for removing apoptotic debris and initiating an innate immune response against pathogens. Despite its importance, the connection between these two functions is poorly understood. Recently it was shown in
C. elegans that mutations in the genes involved in apoptotic cell clearance make them more resistant to the pathogenic bacteria
Pseudomonas aeruginosa and
Salmonella typhimurium [63]. The mutant worms showed an upregulation of the innate immune response genes, even in the absence of bacterial pathogens, and this response was actively regulated by the apoptotic cell clearance defects through the PMK-1 and MPK-1 pathways
[63].
Similar observations were documented in mammals: inefficient removal of dead cells activates the innate immune system
[64]. In DNaseII knockout mice, the DNA of apoptotic cells is not entirely degraded in lysosomes, which leads to the induction of an immune response and the development of chronic arthritis and anaemia
[64]. In line with these data, the production of antibacterial peptides was observed both in
C. elegans and
Drosophila if apoptotic germ cells lacked DNaseII
[65][66]. As there are some controversial data
[67], further investigation is needed to precisely determine the relationship between the innate immune response and apoptotic cell clearance.
Interestingly, recent studies also suggest a neurological connection to immunity, as worms were proven to be able to avoid pathogen attacks through sensing microbes via their nervous system. Multiple sensory neurons and GPCRs are involved in the detection of specific molecules and the local fluctuations of oxygen and carbon dioxide levels generated by bacterial metabolism (reviewed in
[68]). Thus, it is thought that the nervous system can regulate immunity in the nematode, in line with similar findings observed earlier in mammals
[69][70][71].
C. elegans has only 302 neurons with detailed functional and morphological characterization available, and together with the knowledge of the entire synaptic wiring diagram, it can provide useful insights into the research of vertebrate neuroimmunology.