Endocytosis provides the cellular nutrition and homeostasis of organisms, but pathogens often take advantage of this entry point to infect host cells. This is counteracted by phagocytosis that plays a key role in the protection against invading microbes both during the initial engulfment of pathogens and in the clearance of infected cells. Phagocytic cells balance two vital functions: preventing the accumulation of cell corpses to avoid pathological inflammation and autoimmunity, whilst maintaining host defence. In this review, we compare elements of phagocytosis in mammals and the nematode Caenorhabditis elegans. Initial recognition of infection requires different mechanisms. In mammals, pattern recognition receptors bind pathogens directly, whereas activation of the innate immune response in the nematode rather relies on the detection of cellular damage. In contrast, molecules involved in efferocytosis—the engulfment and elimination of dying cells and cell debris—are highly conserved between the two species. Therefore, C. elegans is a powerful model to research mechanisms of the phagocytic machinery. Finally, we show that both mammalian and worm studies help to understand how the two phagocytic functions are interconnected: emerging data suggest the activation of innate immunity as a consequence of defective apoptotic cell clearance.
- endocytosis
- phagocytosis
- apoptosis
- efferocytosis
- pattern recognition receptors
- innate immunity
- Caenorhabditis elegans
1. Forms of Endocytosis
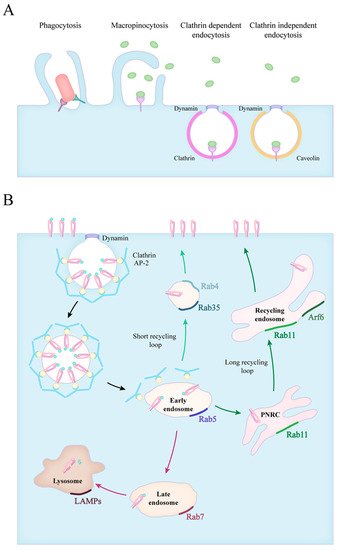
2. Phagocytosis of Apoptotic Cells
2.1. Apoptotic Engulfment in Mammals
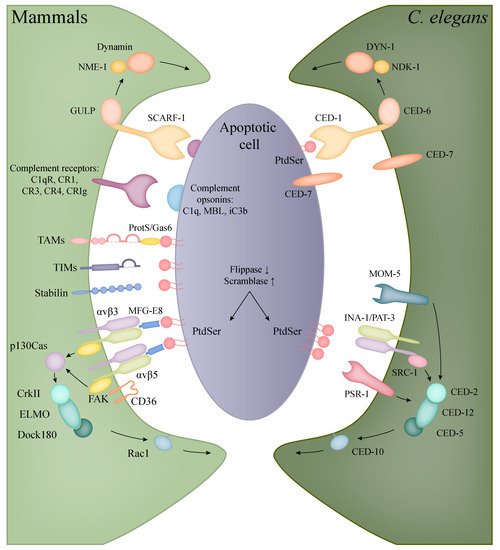
| Human Protein | Function | C. elegans | Protein |
|---|---|---|---|
| ATP8A2 | P4-type ATPase/flippase | TAT-1 | |
| XKR8 | scramblase | CED-8 | |
| SCARF1, MEGF10 and 11, LRP1 (CD91), Jedi-1 | phagocytic receptor | CED-1 | |
| GULP | adaptor | CED-6 | |
| NME1 | nucleoside diphosphate kinase | NDK-1 | |
| Dynamin | large GTPase | DYN-1 | |
| ABCA1 and ABCA7 | ABC transporter | CED-7 | |
| JMJD6 (PSR?) | receptor | PSR-1 | |
| FZD1 and 7 (Frizzled class receptor 1 and 7) | receptor | MOM-5 | |
| integrin α/β chain | receptor | INA-1/PAT-3 | |
| SRC | non-receptor tyrosine kinase | SRC-1 | |
| CrkII | adaptor | CED-2 | |
| ELMO | adaptor | CED-12 | |
| Dock180 | Rac GEF | CED-5 | |
| Rac1 | Rho family GTPase | CED-10 | |
| MFG-E8 | bridging between PtdSer and integrins | - | |
| TIM1, 3, 4 | PtdSer receptor | - | |
| Protein S, GAS6 | bridging between PtdSer and TAM receptors (MER, AXL, TYRO3) | - |
2.2. Apoptotic Engulfment and Clearance in
C. elegans
2.3. The Interconnection of Innate Immunity with Apoptotic Cell Clearance and the Nervous System
References
- Walpole, G.F.W.; Grinstein, S. Endocytosis and the internalization of pathogenic organisms: Focus on phosphoinositides. F1000Research 2020, 9, 1–17.Harald Stenmark; Rab GTPases as coordinators of vesicle traffic. Nature Reviews Molecular Cell Biology 2009, 10, 513-525, 10.1038/nrm2728.
- Cummings, R.J.; Barbet, G.; Bongers, G.; Hartmann, B.M.; Gettler, K.; Muniz, L.; Furtado, G.C.; Cho, J.; Lira, S.A.; Blander, J.M. Different tissue phagocytes sample apoptotic cells to direct distinct homeostasis programs. Nature 2016, 539, 565–569.
- Quintana, J.A.; García-Silva, S.; Mazariegos, M.; González de la Aleja, A.; Nicolás-Ávila, J.A.; Walter, W.; Adrover, J.M.; Crainiciuc, G.; Kuchroo, V.K.; Rothlin, C.V.; et al. Phagocytosis imprints heterogeneity in tissue-resident macrophages. J. Exp. Med. 2017, 214, 1281–1296.
- Roberts, A.W.; Lee, B.L.; Deguine, J.; John, S.; Shlomchik, M.J.; Barton, G.M. Tissue-Resident Macrophages Are Locally Programmed for Silent Clearance of Apoptotic Cells. Immunity 2017, 47, 913–927.e6.
- Fadok, V.A.; Voelker, D.R.; Campbell, P.A.; Cohen, J.J.; Bratton, D.L.; Henson, P.M. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 1992, 148, 2207–2216.
- Andersen, J.P.; Vestergaard, A.L.; Mikkelsen, S.A.; Mogensen, L.S.; Chalat, M.; Molday, R.S. P4-ATPases as Phospholipid Flippases-Structure, Function, and Enigmas. Front. Physiol. 2016, 7, 275.
- Segawa, K.; Kurata, S.; Yanagihashi, Y.; Brummelkamp, T.R.; Matsuda, F.; Nagata, S. Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science 2014, 344, 1164–1168.
- Suzuki, J.; Denning, D.P.; Imanishi, E.; Horvitz, H.R.; Nagata, S. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 2013, 341, 403–406.
- Fadok, V.A.; Bratton, D.L.; Rose, D.M.; Pearson, A.; Ezekewitz, R.A.; Henson, P.M. A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature 2000, 405, 85–90.
- Williamson, P.; Schlegel, R.A. Hide and seek: The secret identity of the phosphatidylserine receptor. J. Biol. 2004, 3, 1–4.
- Klose, R.J.; Kallin, E.M.; Zhang, Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006, 7, 715–727.
- Böse, J.; Gruber, A.D.; Helming, L.; Schiebe, S.; Wegener, I.; Hafner, M.; Beales, M.; Köntgen, F.; Lengeling, A. The phosphatidylserine receptor has essential functions during embryogenesis but not in apoptotic cell removal. J. Biol. 2004, 3, 715–727.
- Umetsu, S.E.; Lee, W.-L.; McIntire, J.J.; Downey, L.; Sanjanwala, B.; Akbari, O.; Berry, G.J.; Nagumo, H.; Freeman, G.J.; Umetsu, D.T.; et al. TIM-1 induces T cell activation and inhibits the development of peripheral tolerance. Nat. Immunol. 2005, 6, 447–454.
- Ichimura, T.; Asseldonk, E.J.P.V.; Humphreys, B.D.; Gunaratnam, L.; Duffield, J.S.; Bonventre, J.V. Kidney injury molecule-1 is a phosphatidylserine receptor that confers a phagocytic phenotype on epithelial cells. J. Clin. Invest. 2008, 118, 1657–1668.
- Monney, L.; Sabatos, C.A.; Gaglia, J.L.; Ryu, A.; Waldner, H.; Chernova, T.; Manning, S.; Greenfield, E.A.; Coyle, A.J.; Sobel, R.A.; et al. Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature 2002, 415, 536–541.
- Anderson, A.C.; Anderson, D.E.; Bregoli, L.; Hastings, W.D.; Kassam, N.; Lei, C.; Chandwaskar, R.; Karman, J.; Su, E.W.; Hirashima, M.; et al. Promotion of tissue inflammation by the immune receptor Tim-3 expressed on innate immune cells. Science 2007, 318, 1141–1143.
- de Mingo Pulido, Á.; Gardner, A.; Hiebler, S.; Soliman, H.; Rugo, H.S.; Krummel, M.F.; Coussens, L.M.; Ruffell, B. TIM-3 Regulates CD103+ Dendritic Cell Function and Response to Chemotherapy in Breast Cancer. Cancer Cell 2018, 33, 60–74.e6.
- Kobayashi, N.; Karisola, P.; Peña-Cruz, V.; Dorfman, D.M.; Jinushi, M.; Umetsu, S.E.; Butte, M.J.; Nagumo, H.; Chernova, I.; Zhu, B.; et al. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity 2007, 27, 927–940.
- Miyanishi, M.; Tada, K.; Koike, M.; Uchiyama, Y.; Kitamura, T.; Nagata, S. Identification of Tim4 as a phosphatidylserine receptor. Nature 2007, 450, 435–439.
- Lemke, G. How macrophages deal with death. Nat. Rev. Immunol. 2019, 19, 539–549.
- Nagata, S. Apoptosis and Clearance of Apoptotic Cells. Annu. Rev. Immunol. 2018, 36, 489–517.
- Lew, E.D.; Oh, J.; Burrola, P.G.; Lax, I.; Zagórska, A.; Través, P.G.; Schlessinger, J.; Lemke, G. Differential TAM receptor-ligand-phospholipid interactions delimit differential TAM bioactivities. Elife 2014, 3, 1–23.
- Seitz, H.M.; Camenisch, T.D.; Lemke, G.; Earp, H.S.; Matsushima, G.K. Macrophages and dendritic cells use different Axl/Mertk/Tyro3 receptors in clearance of apoptotic cells. J. Immunol. 2007, 178, 5635–5642.
- Rothlin, C.V.; Ghosh, S.; Zuniga, E.I.; Oldstone, M.B.A.; Lemke, G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell 2007, 131, 1124–1136.
- van der Meer, J.H.M.; van der Poll, T.; van ’t Veer, C. TAM receptors, Gas6, and protein S: Roles in inflammation and hemostasis. Blood 2014, 123, 2460–2469.
- Hanayama, R.; Tanaka, M.; Miwa, K.; Shinohara, A.; Iwamatsu, A.; Nagata, S. Identification of a factor that links apoptotic cells to phagocytes. Nature 2002, 417, 182–187.
- Stern, M.; Savill, J.; Haslett, C. Human monocyte-derived macrophage phagocytosis of senescent eosinophils undergoing apoptosis. Mediation by alpha v beta 3/CD36/thrombospondin recognition mechanism and lack of phlogistic response. Am. J. Pathol. 1996, 149, 911–921.
- Albert, M.L.; Pearce, S.F.; Francisco, L.M.; Sauter, B.; Roy, P.; Silverstein, R.L.; Bhardwaj, N. Immature dendritic cells phagocytose apoptotic cells via alphavbeta5 and CD36, and cross-present antigens to cytotoxic T lymphocytes. J. Exp. Med. 1998, 188, 1359–1368.
- Savill, J.; Hogg, N.; Ren, Y.; Haslett, C. Thrombospondin cooperates with CD36 and the vitronectin receptor in macrophage recognition of neutrophils undergoing apoptosis. J. Clin. Invest. 1992, 90, 1513–1522.
- Albert, M.L.; Kim, J.I.; Birge, R.B. alphavbeta5 integrin recruits the CrkII-Dock180-rac1 complex for phagocytosis of apoptotic cells. Nat. Cell Biol. 2000, 2, 899–905.
- Gumienny, T.L.; Brugnera, E.; Tosello-Trampont, A.C.; Kinchen, J.M.; Haney, L.B.; Nishiwaki, K.; Walk, S.F.; Nemergut, M.E.; Macara, I.G.; Francis, R.; et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 2001, 107, 27–41.
- Shah, P.P.; Fong, M.Y.; Kakar, S.S. PTTG induces EMT through integrin αVβ3-focal adhesion kinase signaling in lung cancer cells. Oncogene 2012, 31, 3124–3135.
- Liang, Y.Y.; Arnold, T.; Michlmayr, A.; Rainprecht, D.; Perticevic, B.; Spittler, A.; Oehler, R. Serum-dependent processing of late apoptotic cells for enhanced efferocytosis. Cell Death Dis. 2014, 5, e1264.
- Nauta, A.J.; Castellano, G.; Xu, W.; Woltman, A.M.; Borrias, M.C.; Daha, M.R.; van Kooten, C.; Roos, A. Opsonization with C1q and mannose-binding lectin targets apoptotic cells to dendritic cells. J. Immunol. 2004, 173, 3044–3050.
- Fraser, D.A.; Laust, A.K.; Nelson, E.L.; Tenner, A.J. C1q differentially modulates phagocytosis and cytokine responses during ingestion of apoptotic cells by human monocytes, macrophages, and dendritic cells. J. Immunol. 2009, 183, 6175–6185.
- Takizawa, F.; Tsuji, S.; Nagasawa, S. Enhancement of macrophage phagocytosis upon iC3b deposition on apoptotic cells. FEBS Lett. 1996, 397, 269–272.
- Matsui, H.; Tsuji, S.; Nishimura, H.; Nagasawa, S. Activation of the alternative pathway of complement by apoptotic Jurkat cells. FEBS Lett. 1994, 351, 419–422.
- Mevorach, D.; Mascarenhas, J.O.; Gershov, D.; Elkon, K.B. Complement-dependent clearance of apoptotic cells by human macrophages. J. Exp. Med. 1998, 188, 2313–2320.
- Benoit, M.E.; Clarke, E.V.; Morgado, P.; Fraser, D.A.; Tenner, A.J. Complement protein C1q directs macrophage polarization and limits inflammasome activity during the uptake of apoptotic cells. J. Immunol. 2012, 188, 5682–5693.
- Fraser, D.A.; Arora, M.; Bohlson, S.S.; Lozano, E.; Tenner, A.J. Generation of inhibitory NFkappaB complexes and phosphorylated cAMP response element-binding protein correlates with the anti-inflammatory activity of complement protein C1q in human monocytes. J. Biol. Chem. 2007, 282, 7360–7367.
- Conradt, B.; Xue, D. Programmed Cell Death; WormBook: Pasadena, CA, USA, 2005; pp. 1–13.
- Conradt, B.; Wu, Y.-C.; Xue, D. Programmed Cell Death During Caenorhabditis elegans Development. Genetics 2016, 203, 1533–1562.
- Bratton, D.L.; Fadok, V.A.; Richter, D.A.; Kailey, J.M.; Guthrie, L.A.; Henson, P.M. Appearance of phosphatidylserine on apoptotic cells requires calcium-mediated nonspecific flip-flop and is enhanced by loss of the aminophospholipid translocase. J. Biol. Chem. 1997, 272, 26159–26165.
- Lu, N.; Zhou, Z. Membrane trafficking and phagosome maturation during the clearance of apoptotic cells. Int. Rev. Cell Mol. Biol. 2012, 293, 269–309.
- Mangahas, P.M.; Zhou, Z. Clearance of apoptotic cells in Caenorhabditis elegans. Semin. Cell Dev. Biol. 2005, 16, 295–306.
- Kinchen, J.M.; Cabello, J.; Klingele, D.; Wong, K.; Feichtinger, R.; Schnabel, H.; Schnabel, R.; Hengartner, M.O. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature 2005, 434, 93–99.
- Shen, Q.; He, B.; Lu, N.; Conradt, B.; Grant, B.D.; Zhou, Z. Phagocytic receptor signaling regulates clathrin and epsin-mediated cytoskeletal remodeling during apoptotic cell engulfment in C. elegans. Development 2013, 140, 3230–3243.
- Zhou, Z.; Hartwieg, E.; Horvitz, H.R. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 2001, 104, 43–56.
- Venegas, V.; Zhou, Z. Two alternative mechanisms that regulate the presentation of apoptotic cell engulfment signal in Caenorhabditis elegans. Mol. Biol. Cell 2007, 18, 3180–3192.
- Yu, X.; Odera, S.; Chuang, C.-H.; Lu, N.; Zhou, Z. C. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev. Cell 2006, 10, 743–757.
- Schmid, S.L.; Frolov, V.A. Dynamin: Functional design of a membrane fission catalyst. Annu. Rev. Cell Dev. Biol. 2011, 27, 79–105.
- Farkas, Z.; Petric, M.; Liu, X.; Herit, F.; Rajnavölgyi, É.; Szondy, Z.; Budai, Z.; Orbán, T.I.; Sándor, S.; Mehta, A.; et al. The nucleoside diphosphate kinase NDK-1/NME1 promotes phagocytosis in concert with DYN-1/Dynamin. FASEB J. 2019, 33, 11606–11614.
- Yu, X.; Lu, N.; Zhou, Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 2008, 6, e61.
- Reddien, P.W.; Horvitz, H.R. The engulfment process of programmed cell death in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 2004, 20, 193–221.
- Hsu, T.-Y.; Wu, Y.-C. Engulfment of apoptotic cells in C. elegans is mediated by integrin alpha/SRC signaling. Curr. Biol. 2010, 20, 477–486.
- Wang, X.; Wu, Y.-C.; Fadok, V.A.; Lee, M.-C.; Gengyo-Ando, K.; Cheng, L.-C.; Ledwich, D.; Hsu, P.-K.; Chen, J.-Y.; Chou, B.-K.; et al. Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science 2003, 302, 1563–1566.
- Hsieh, H.-H.; Hsu, T.-Y.; Jiang, H.-S.; Wu, Y.-C. Integrin α PAT-2/CDC-42 signaling is required for muscle-mediated clearance of apoptotic cells in Caenorhabditis elegans. PLoS Genet. 2012, 8, e1002663.
- Neukomm, L.J.; Zeng, S.; Frei, A.P.; Huegli, P.A.; Hengartner, M.O. Small GTPase CDC-42 promotes apoptotic cell corpse clearance in response to PAT-2 and CED-1 in C. elegans. Cell Death Differ. 2014, 21, 845–853.
- Cabello, J.; Neukomm, L.J.; Günesdogan, U.; Burkart, K.; Charette, S.J.; Lochnit, G.; Hengartner, M.O.; Schnabel, R. The Wnt pathway controls cell death engulfment, spindle orientation, and migration through CED-10/Rac. PLoS Biol. 2010, 8, e1000297.
- Klinkert, K.; Echard, A. Rab35 GTPase: A Central Regulator of Phosphoinositides and F-actin in Endocytic Recycling and Beyond. Traffic 2016, 17, 1063–1077.
- Haley, R.; Wang, Y.; Zhou, Z. The small GTPase RAB-35 defines a third pathway that is required for the recognition and degradation of apoptotic cells. PLoS Genet. 2018, 14, e1007558.
- Wan, J.; Yuan, L.; Jing, H.; Zheng, Q.; Xiao, H. Defective apoptotic cell clearance activates innate immune response to protect Caenorhabditis elegans against pathogenic bacteria. Virulence 2021, 12, 75–83.
- Kawane, K.; Ohtani, M.; Miwa, K.; Kizawa, T.; Kanbara, Y.; Yoshioka, Y.; Yoshikawa, H.; Nagata, S. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature 2006, 443, 998–1002.
- Yu, H.; Lai, H.-J.; Lin, T.-W.; Chen, C.-S.; Lo, S.J. Loss of DNase II function in the gonad is associated with a higher expression of antimicrobial genes in Caenorhabditis elegans. Biochem. J. 2015, 470, 145–154.
- Mukae, N.; Yokoyama, H.; Yokokura, T.; Sakoyama, Y.; Nagata, S. Activation of the innate immunity in Drosophila by endogenous chromosomal DNA that escaped apoptotic degradation. Genes Dev. 2002, 16, 2662–2671.
- Seong, C.-S.; Varela-Ramirez, A.; Aguilera, R.J. DNase II deficiency impairs innate immune function in Drosophila. Cell. Immunol. 2006, 240, 5–13.
- Liu, Y.; Sun, J. Detection of Pathogens and Regulation of Immunity by the Caenorhabditis elegans Nervous System. MBio 2021, 12, 1–12.
- Sternberg, E.M. Neural regulation of innate immunity: A coordinated nonspecific host response to pathogens. Nat. Rev. Immunol. 2006, 6, 318–328.
- Steinman, L. Elaborate interactions between the immune and nervous systems. Nat. Immunol. 2004, 5, 575–581.
- Tracey, K.J. The inflammatory reflex. Nature 2002, 420, 853–859.
