
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Liang WU | + 3233 word(s) | 3233 | 2021-08-28 07:31:12 | | | |
| 2 | Conner Chen | + 720 word(s) | 3953 | 2021-09-23 02:48:58 | | |
Video Upload Options
The precise segmentation of brain tumor images is a vital step towards accurate diagnosis and effective treatment of brain tumors. Magnetic Resonance Imaging (MRI) can generate brain images without tissue damage or skull artifacts, providing important discriminant information for clinicians in the study of brain tumors and other brain diseases. Segmentation methods of brain tumor MR image are mainly divided into three categories according to different segmentation principles: traditional segmentation methods, traditional machine learning-based segmentation methods and deep learning-based segmentation methods.
1. Traditional Brain Tumor Segmentation Methods
1.1. Segmentation Methods Based on Threshold
Threshold-based segmentation is the simplest method. First, it is assumed that the pixels within a range belong to the same category [1]. Brain tumor images can be divided into target region and background region by setting an appropriate threshold. Different thresholds can also be set to divide the tumor into multiple regions. After continuous research and development, the accuracy of threshold segmentation has been greatly improved. Wang Y P et al. proposed an improved threshold segmentation algorithm. The method improves the noise sensitivity in threshold segmentation by using local information of pixel neighborhood [2]. Foladivanda et al. proposed an adaptive threshold segmentation method. The method can effectively overcome the problem of uneven gray, and enhance the contrast of images, and effectively improve the DSC and JS measure of MR image segmentation of the brain tumor [3].
The segmentation method based on threshold is relatively simple, and the quality of segmentation results almost entirely depends on the size of threshold, so the selection of threshold is very important. Moreover, the threshold segmentation method can only segment simple images, and it is difficult to deal with complex images.
1.2. Segmentation Methods Based on Region
Common region-based segmentation methods include watershed algorithm and region-growing algorithm.
Watershed algorithm is a segmentation method based on mathematical morphology. In this algorithm, the image to be processed is compared to the terrain in geography, and the elevation of terrain is represented by the gray value of the pixel. The local minimum and its adjacent area are called the ponding basin. It is assumed that there are water permeable holes at each local minimum. With the increase of infiltration water, the ponding basin will be gradually submerged. Blocking the flow of water from a stagnant basin to a nearby basin is called a dam. When the water level reaches the peak, the infiltration process ends. These dams are called watersheds. Kaleem et al. [4] proposed a watershed segmentation method guided by setting internal or external markers to calculate the morphological gradient of the input image and internal and external markers of the original image. Then they use watershed transform to obtain the segmentation results. Rajini N et al. [5] proposed a method combining threshold segmentation and watershed. First, the image was segmented by threshold method, and then the segmented image was segmented by watershed algorithm. The experiment proved that the segmentation results obtained by this method were more accurate than those obtained by one of the two methods alone, with the average TPR measure higher than 90%.
The segmentation algorithm based on watershed can obtain a complete closed curve and provide contour information for subsequent processing, whereas the watershed algorithm is influenced by noise and easy to over segment.
The region growing algorithm draws all the pixel points conforming to the criterion into the same region via formulating a criterion, so as to achieve pixel segmentation. This kind of segmentation method has the following characteristics: (1) Each pixel must be in a certain region, and the pixels in the region must be connected, and must meet certain similar conditions; (2) different regions are disjoint, and two different regions cannot have the same property. Qusay et al. [6] proposed an automatic seed region growth method, which can automatically set the initial value of seeds, avoid the defects of manual interaction, and improve the efficiency of image segmentation.
The region-based segmentation method has the characteristics of simple calculation and high accuracy, which can extract better regional features and is more suitable for segmentation of small targets. However, it is sensitive to noise and easy to make holes in the extracted region.
1.3. Segmentation Methods Based on Fuzzy Theory
The segmentation methods based on fuzzy theory have also been highly valued. In brain tumor MR image segmentation, the most widely used Fuzzy theory algorithm is Fuzzy C-means clustering (FCM) [7]. Muneer K et al. [8] obtained the K-FCM method through the combination of FCM algorithm and K-means algorithm. The experiment proved that, compared with FCM, K-FCM showed higher accuracy in brain tumor MR image segmentation and could reduce the computational complexity. Guo Y et al. [9] proposed a Neutrosophic C-Means (NCM) algorithm based on fuzzy C-means and neutral set framework. The algorithm introduced distance constraint into the objective function to solve the problem of insufficient prior knowledge and achieved satisfactory segmentation results. On the basis of Super-pixel fuzzy clustering and the lattice Boltzmann method, Asieh et al. [10] proposed a level set method that can automatically segment brain tumors, which has strong robustness to image intensity and noise.
The segmentation method based on fuzzy theory can effectively solve the problem of incomplete image information, imprecision, and so on. It has strong compatibility and can be used in combination with other methods, but it is difficult to deal with large-scale data due to its large amount of computation and high time complexity.
1.4. Segmentation Methods Based on Edge Detection
The segmentation principle based on edge detection and target contour achieves segmentation by obtaining the edge of the target region and then obtaining the contour of the target region. Common detection operators for edge detection include Roberts operator, Sobel operator, Canny operator and Prewitt operator [11]. Jayanthi et al. [12] integrated FCM into the active contour model. The initial contour of the model is automatically selected by FCM, which reduces the human–computer interaction. Moreover, the problem of the unclear edge contour and uneven intensity in MR images was improved. The average DSC measure of segmentation by this method reached 81%.
Compared with other traditional segmentation methods, the segmentation method based on edge detection pays attention to the edge information of the image and links the edges into contours, and the anti-noise performance is stronger. But the anti-noise performance is negatively correlated with accuracy, that is, the better the anti-noise performance, the lower the accuracy. On the contrary, improved accuracy will reduce the anti-noise performance.
2. Segmentation Methods of Brain Tumor MR Images Based on Traditional Machine Learning
Brain tumor segmentation methods based on traditional machine learning use predefined features to train the classification model. Generally, they are divided into two levels: organizational level and pixel level. At the organizational level, the classifier needs to determine which kind of organizational structure each feature belongs to, and at the pixel level the classifier needs to determine which category each pixel belongs to. Traditional Machine Learning algorithms mainly include K-Nearest Neighbors (KNN) [13], Support Vector Machine (SVM) [14], Random Forest (RF) [15], Dictionary Learning (DL) [16], etc.
Havaei et al. [13] regarded each brain as a separate database and used the KNN algorithm for segmentation. They obtained very accurate results, and the segmentation time of each brain image is only one minute, which improves the efficiency of segmentation. Llner F et al. [14] used SVM to segment brain tumors, taking into account the changing characteristics of signal intensity and other features of brain tumor MR images. The TPR measure of this method for LGG reached 83%, and the accuracy measure for HGG reached 91%. Sher et al. [17] first segmented the image by the Otsu method and K-means clustering, then extracted the features by discrete wavelet transformation, and finally reduced the feature dimension by the PCA algorithm to obtain the best features for SVM classification. The experimental results show that the sensitivity and specificity of the scheme can reach more than 90%. Vaishnavee et al. [18] used a proximal support vector machine (PSVM). The method uses equation constraints to solve the primary linear equations, which simplifies the original problem of solving convex quadratic programming. The experiment shows that PSVM is more accurate than SVM in MR image segmentation of brain tumor. Wu et al. [19] proposed a method to first segment the image into super-voxels, then segment the tumor using MRF, estimate the likelihood function at the same time, and extract the features using a multistage wavelet filter. Nabizadeh et al. [20] proposed an automatic segmentation algorithm based on texture and contour. Firstly, the initial points were determined and the machine learning classifier was trained by the initial points. Mahmood et al. [15] proposed an automatic segmentation algorithm based on RF. This algorithm uses several important features such as image intensity, gradient and entropy to generate multiple classifiers, and classifies pixels in multispectral brain MR images by combining the results to obtain segmentation results. Selvathi et al. [21] increased the weight of the wrongly classified samples and decreased the weight of the correctly classified samples in the training process. Then the classifier gives new weights to the samples to ensure that the weights of all decision trees are positively correlated with their classification ability. Finally, the input of the improved RF consists of two parts: the image intensity feature and the original image feature extracted by curve and wavelet transformation. Experimental results show that the accuracy of the improved RF scheme is 3% higher than that of the original RF algorithm. Reza et al. [22] studied the correlation of image minimization features from the perspective of image features, effectively selected features, and finally classified features in multimodal MR images through RF. Compared with the RF algorithm alone, the proposed method can improve the DSC, PPV and TPR measure simultaneously. Meier et al. [23] trained a specific random forest classifier by semi-supervised learning. It takes image segmentation as a classification task and effectively combines the preoperative and postoperative MR image information to improve the postoperative brain tumor segmentation. The PPV and ME measure obtained by this method were 93% and 2.4%, respectively. Dictionary learning is a kind of learning method for simulating dictionary lookup. The dictionary itself is set as dictionary matrix, and the method used is sparse matrix. The process of dictionary lookup is obtained by multiplying the sparse matrix and dictionary matrix, and then the dictionary matrix and sparse matrix are optimized to minimize the error between the value searched and the original data. Chen et al. [16] transformed the super-pixel feature into a high-dimensional feature space. According to the different error values of different regions when the dictionary was modeling brain tumors, the segmentation of brain tumor MR images was realized and the segmentation accuracy was improved. Li [24] proposed a multi dictionary fuzzy learning algorithm based on dictionary learning. This algorithm effectively combines dictionary learning with fuzzy algorithm, and fully considers the differences between the target region and the background, as well as the consistency within the target region. This method can describe the gray and texture information of different regions of the image, and segment the image quickly and accurately.
The traditional machine learning algorithm is better than many traditional segmentation algorithms in algorithmic performance, but there are many shortcomings when it is used in brain tumor MR image segmentation. For example, the KNN algorithm is simple to implement, and the prediction accuracy of the brain tumor region is relatively high, but the calculation is relatively large [13]. The support vector machine has strong theory, and the final result is determined by several support vectors. The calculation is relatively simple and the generalization ability is strong, but it has higher requirements concerning the selection of parameters and kernel function [14]. Random forest can solve the problem of over-segmentation well, process multiple types of data, and has good anti-noise performance. It can parallel operation and shorten the operation time, but it has a poor effect on low-dimensional tumor data processing [15]. The algorithm based on dictionary learning is similar to the idea of dimensionality reduction, both of which reduce the computing complexity and speed up the computing speed, but also have higher requirements for tumor data [16].
3. Segmentation Methods of Brain Tumor MR Images Based on Deep Learning
3.1. Segmentation Methods of Brain Tumor MR Images Based on CNN
Convolutional neural network belongs to the category of neural network, and its weight sharing mechanism greatly reduces the model complexity. Convolutional neural network (the network diagram is shown in Figure 1a) can directly take the image as the input, automatically extract the features, and has a high degree of invariance to the image translation, scaling and other changes. In recent years, a series of Network models based on convolutional neural Network [25], such as Network in Network [26], VGG [27], Google-Net [28], Res-Net [29], etc., have been widely used in medical image segmentation. Among them, the VGG network has a strong ability to extract features and can guarantee the convergence in the case of fewer training times. However, as the deepening of the network will cause gradient explosion and gradient disappearance, the optimization effect will start to deteriorate when the network depth exceeds a certain range.
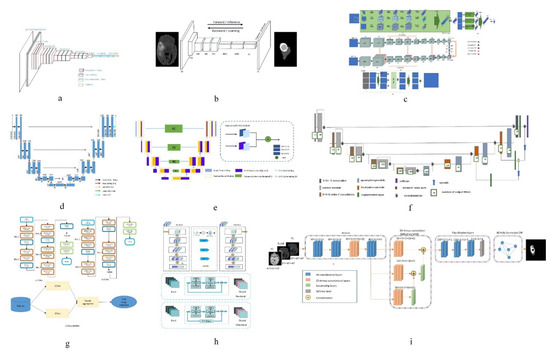
Figure 1. Network structure diagrams of some classical methods and improved methods. (a) is the classic CNN network model; (b) is the classic FCN network model; (c) is the structure diagram of a convolutional neural network technology based on automatic context (Auto-Nets) proposed by Salehi et al. [30]; (d) is the classic Encoder-Decoder network model; (e) is the structure diagram of a fully convolutional neural network with feature reuse module and feature integration module (f2fcn) proposed by Xue et al. [31]; (f) is the structure diagram of a robust neural network algorithm based on u-net proposed by isensee et al. [32]; (g) is the structure diagram of a sequential CNN architecture proposed by saouli et al. [33]; (h) is the structure diagram of attention residual U-net proposed by Zhang et al. [34]; (i) is a structural diagram of 3D dense connection combined with feature pyramid proposed by Zhou et al. [35].
In order to solve the problem of network degradation, He et al. [29] proposed deep Residual Network (ResNet), which achieved good results in the segmentation task [36]; Anand et al. [37] combined the 3D convolutional neural network with dense connection, pre-trained the model, and then initialized the model with the weight obtained. This method improved the DSC measure in the segmentation task of brain tumor MR images. Havaei et al. [38] constructed a cascaded dual path CNN, which took the output characteristic graph of CNN in the first stage as the additional input of CNN in the second stage. This method can effectively obtain rich background information and get better segmentation results. Lai et al. [39] reduced the tail of the original image by 98% firstly, corrected the bias field by using n4itk, then pre-segmented it by multi classification CNN, and finally obtained the final segmentation result by median filtering. The algorithm improves the DSC and PPV of segmentation significantly. Salehi et al. [30] proposed a convolutional neural network technology based on automatic context (Auto-Nets) to indirectly learn 3D image information by means of 2D convolution. This method uses 2D convolution in axial, coronal and sagittal MR images respectively to avoid complex 3D convolution operations in segmentation (The network diagram is shown in Figure 1c). Hussain et al. [40] established a correlation architecture composed of a parallel CNN layer and a linear CNN layer by adding an induction structure. This structure has achieved good results in brain tumor MR image segmentation, especially in enhancing the DSC measure to 90%. Kamnitsas et al. [41] trained 3D brain tumor images and then carried out conditional random field post-processing to obtain smoother results. Saouli et al. [33] designed a sequential CNN architecture and proposed that an end-to-end incremental network can simultaneously develop and train CNN models (the network diagram is shown in Figure 1g). The average DSC measure obtained by this method is 88%. Hu K et al. [42] proposed a more hierarchical convolution based Neural Network (Multi-Cascaded Convolutional Neural Network, MCCNN) and fully connected conditional random fields (CRFs), combined with the brain tumor segmentation method, Firstly, the brain tumor is roughly segmented by multi classification convolution neural network, and then fine segmented by fully connected random field according to the rough segmentation results, so as to achieve the effect of batch segmentation and improve the accuracy. The segmentation algorithm based on CNN can automatically extract features and process high-dimensional data, but it is easy to lose information in the process of pooling, and its interpretability is poor.
3.2. Segmentation Methods of Brain Tumor MR Images Based on FCN
Compared with pixel-level classification, image-level classification and regression tasks are more suitable for using the CNN structure, because they both expect to obtain a probable value for image classification. For semantic segmentation of images, FCN works better. FCN has no requirement on the size of the input image, and there will be an up sampling process at the last convolution layer. This process can get the same result as the input image size, predicting each pixel while retaining the spatial information in the input image, so as to achieve the pixel classification. In simple terms, FCN is a method to classify and segment images at the pixel level. Therefore, the semantic segmentation model based on FCN is more in line with the requirements of medical image segmentation. Zhao et al. [43] proposed a combination of FCN with CRF for brain tumor segmentation. The method trains two-dimensional slices in axial, coronal and sagittal directions respectively, and then uses fusion strategy to combine segmented brain tumor images. Compared with the traditional segmentation methods, the segmentation speed is faster and the efficiency is higher. Xue et al. [31] proposed a fully convolutional neural network with feature reuse module and feature integration module (f2fcn). It reuses the features of different layers, and uses the feature integration module to eliminate the possible noise and enhance the fusion between different layers (the network diagram is shown in Figure 1e). The DSC and PPV obtained by this method are high. Zhou et al. [44] proposed a 3D atomic convolution feature pyramid to enhance the discrimination ability of the model, which is used to segment tumors of different sizes. Then, an improvement is made on the original basis [35], a 3D dense connection architecture is proposed, and a new feature pyramid module is designed by using 3D convolution (the network diagram is shown in Figure 1i). This module is used to fuse multi-scale context to improve the accuracy of segmentation. Liu et al. [45] proposed a Dilated Convolution optimization structure (DCR) based on Resnet-50, which can effectively extract local and global features, and this method can improve the segmentation PPV measure to 92%. The segmentation algorithm based on FCN can predict the category of each pixel, transform the image classification level to the semantic level, retain the position information in the original image, and obtain a result with the same size as the input image. However, the algorithm has low computational efficiency, takes up a lot of memory space, and the receptive field is relatively small.
3.3. Segmentation Methods of Brain Tumor MR Images Based on Encoder-Decoder Structure
The encoder-decoder structure is generally composed of an encoder and a decoder. The encoder trains and learns the input image through a neural network to obtain its characteristic map. The function of the decoder is to mark the category of each pixel after the encoder provides the feature map, so as to achieve the segmentation effect. In the segmentation tasks based on encoder-decoder structure, the structure of encoders is generally similar, mostly derived from the network structure of classification tasks, such as VGG, etc. The purpose of doing this is to obtain the weight parameters of network training through the training of a large database. Therefore, the difference of the decoder reflects the difference of the whole network to a large extent, and is also the key factor affecting the segmentation effect.
Badrinarayanan et al. [46] proposed the SegNet model. Compared with other models, this model has a deeper layer and has better performance in semantic segmentation of pixels. The encoder part of the model consists of a 13 layer vgg-16 network, and can remember the position information of the largest pixel in the encoding phase. In the decoder, the low resolution input features are up sampled to get the segmentation results. The U-Net model based on FCN is a kind of widely used brain tumor segmentation model, in which the network structure is also made up of an encoder and a decoder, and a U-Net network jump connection will code paths, used to get the characteristics of the figure to the decoding path to the corresponding position, in order to get the characteristics of the direct sampling under the coding phase into the decoding stage, thus learning more detailed characteristics. Chen et al. [47] proposed a multi-level deep network, which can obtain image multi-level information by adding auxiliary classifiers on Multi-Level Deep Medical (MLDM) and U-Net, so as to realize image segmentation. The results of DSC, PPV and TPR were 83%, 73% and 85%, respectively. In order to reduce the semantic gap between the feature mapping of encoder and decoder networks, Zhou et al. [48] proposed a variety of nested dense connection methods to connect the encoder and decoder networks. Alom et al. [49] proposed a recursive neural network and a recursive residual convolutional neural network based on U-Net. The experimental results show that the performance of the two kinds of network segmentation combined with U-Net is better than that of U-Net alone. Zhang et al. [34] introduced the attention mechanism and residual network into the traditional U-Net network and proposed an attention residual U-Net (the network diagram is shown in Figure 5h), which improved the segmentation performance of brain tumor MR images. Milletari et al. [50] proposed the V-Net model on the basis of the 3D U-Net model, which extended the original U-Net model by using a 3D convolution check. Hua et al. [51] cascaded V-Net and used the method of segmentation of the whole tumor first into sub-regions of the tumor; the accuracy of segmentation is higher than that of direct V-Net segmentation. Cicek et al. [52] proposed a 3D U-Net model to learn the features of sparse annotated volume images. On the basis of 3D U-Net, Heet et al. [53] added a Hybrid Dilated Convolution (HDC) module to increase the sensory field of neurons, overcoming the restriction that multi-scale feature extraction requires deep neural networks. Using shallow neural networks can reduce the number of model parameters and reduce the computational complexity. Tsenget et al. [54] proposed one with the depth of the layer cross-modal convolution encoder/decoder structure, in combination with MR image data of different modalities, and at the same time using the weighted and multi-stage training methods to solve the problem of unbalanced data; compared with the traditional U-Net structure, the methods of DSC, TPR and PPV measure are improved. Isensee et al. [32] improved the U-Net network model and designed a robust neural network algorithm, which prevented overfitting by expanding the amount of data (the network diagram is shown in Figure 5f). This algorithm improved the TPR measure to 91%; Haichun et al. [55] cleverly applied the improved full convolutional neural network structure to the U-Net model and proposed a novel end-to-end brain tumor segmentation method. In this method, an up-hop connection structure was designed between the encoding path and decoding path to enhance the information flow. Jia et al. [56] constructed a HNF network based on the parallel multi-scale fusion (PMF) module, and proposed a three-dimensional high-resolution and non-local feature network (HNF-NET) for multi parameter MR imaging, which can generate strong high-resolution feature representation and aggregate multi-scale context information. The expectation maximization attention (EMA) module is introduced to extract more relevant features and reduce redundant features. The DSC and HD of the whole tumor are 91.1% and 4.13%, respectively. The segmentation algorithm based on encoder-decoder can combine high-resolution and low-resolution information, and can recognize features from multiple scales, but there is only a short connection between the encoding process and the decoding process, and the connection between the two is obviously insufficient.
References
- Sujan, M.; Alam, N.; Abdullah, S.; Islam, M.J. A Segmentation Based Automated System for Brain Tumor Detection. Comput. Appl. 2016, 153, 41–49.
- Wang, Y.P. Medical Image Processing; Tsinghua University Press: Beijing, China, 2012.
- Fooladivanda, A.; Shokouhi, S.B.; Ahmadinejad, N.; Mosavi, M.R. Automatic Segmentation of Breast and Fibro glandular Tissue in Breast MRI Using Local Adaptive Thresholding. In Proceedings of the 2014 21th Iranian Conference on Biomedical Engineering, ICBME, Tehran, Iran, 26–28 November 2014; pp. 195–200.
- Kaleem, M.; Sanaullah, M.; Hussain, M.A.; Jaffar, M.A.; Choi, T.S. Segmentation of Brain Tumor Tissue Using Marker Controlled Watershed Transform Method. Commun. Comput. Inf. Sci. 2012, 281, 222–227.
- Rajini, N.; Narmatha, T.; Bhavani, R. Automatic Classification of MR Brain Tumor Images Using Decision Tree. In Proceedings of the International Conference on Electronics, Communication and Information Systems, Near Madurai, Tamilnadu, India, 2–3 November 2012; pp. 10–13.
- Qusay, A.; Isa, N. Computer-aided Segmentation System for Breast MRI Tumor Using Modified Automatic Seeded Region Growing. BMRI-MASRG. J. Digit. Imaging 2014, 27, 133–144.
- Lei, T.; Zhang, X.; Jia, X.H. Research Progress of Image Segmentation Based on Fuzzy Clustering. Chin. J. Electron. 2019, 47, 1776–1791.
- Muneer, K.; Joseph, K. Performance Analysis of Combined K-mean and Fuzzy-c-Mean Segmentation of MR Brain Images. Computational Vision and Bio Inspired Computing; Springer: Cham, Switzerland; Berlin/Heidelberg, Germany, 2018; pp. 830–860.
- Guo, Y.; Sengur, A. NCM: Neutrosophic C-Means Clustering Algorithm. Pattern Recognit. 2015, 48, 2710–2724.
- Khosravanian, A. Fast Level Set Method for Glioma Brain Tumor Segmentation Based on Super Pixel Fuzzy Clustering and Lattice Boltzmann Method. Comput. Methods Programs Biomed. 2020, 198, 105809.
- Canny, A. Computational Approach to Edge Detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986, 8, 679–698.
- Jayanthi, S.; Ranganathan, H.; Palanivelan, M. Segmenting Brain Tumor Regions with Fuzzy Integrated Active Contours. IETE J. Res. 2019.
- Havaei, M.; Jodoin, P.M.; Larochelle, A.H. Efficient Interactive Brain Tumor Segmentation as Within-Brain KNN Classification. In Proceedings of the International Conference on Pattern Recognition, Stockholm, Sweden, 24–28 August 2014; pp. 556–561.
- Llner, F.; Emblem, K.; Schad, L. Support Vector Machines in DSC-based Glioma Imaging: Suggestions for Optimal Characterization. Magn. Reson. Med. 2010, 64, 1230–1236.
- Mahmood, Q.; Basit, A. Automatic Ischemic Stroke Lesion Segmentation in Multi-Spectral MRI Images Using Random Forests Classifier; Springer: New York, NY, USA, 2015; pp. 266–274.
- Chen, X.; Binh, P. Automated Brain Tumor Segmentation Using Kernel Dictionary Learning and Super Pixel-level Features. Syst. Man Cybern. 2016, 10, 1109.
- Shil, S.; Polly, F. An Improved Brain Tumor Detection and Classification Mechanism. In Proceedings of the 2017 International Conference on Information and Communication Technology Convergence, ICTC, Jeju, Korea, 18–20 October 2017; pp. 54–57.
- Vaishnavee, K.; Amshakala, K. An Automated MRI Brain Image Segmentation and Tumor Detection Using SOM-clustering and Proximal Support Vector Machine Classifier. In Proceedings of the 2015 IEEE International Conference on Engineering and Technology, ICETECH, Coimbatore, India, 20–20 March 2015; pp. 1–6.
- Wu, W.; Chen, A. Brain Tumor Detection and Segmentation in A CRF (Conditional Random Fields) Framework with Pixel-pairwise Affinity and Super Pixel-level Features. Int. J. Comput. Assist. Radiol. Surg. 2014, 9, 241–253.
- Nabizadeh, N.; Kubat, M. Automatic Tumor Segmentation in Single-spectral MRI Using A Texture-based and Contour-based Algorithm. Expert Syst. Appl. 2017, 77, 1–10.
- Selvathi, D.; Selvaraj, H. Segmentation of Brain Tumor Tissues in MR Images Using Multiresolution Transforms and Random Forest Classifier with Ada Boost Technique. In Proceedings of the 2018 26th International Conference on Systems Engineering, ICSEng, Sydney, Australia, 18–20 December 2018; pp. 1–7.
- Reza, S. Multi-fractal Texture Features for Brain Tumor and Edema Segmentation. In Medical Imaging 2014: Computer-Aided Diagnosis; International Society for Optics and Photonics: San Diego, CA, USA, 2014; Volume 9035, p. 903503.
- Meier, R.; Bauer, S. Patient-specific Semi-supervised Learning for Postoperative Brain Tumor Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; Springer: Cham, Switzerland; Berlin/Heidelberg, Germany, 2014; pp. 714–721.
- Li, Y.F. A Fuzzy Method for Image Segmentation Based on Multi-dictionary Learning. Chin. J. Electron. 2018, 46, 1700–1709.
- Chen, S.H.; Liu, W.X.; Qin, J.; Chen, L.; Bin, G.; Zhou, Y.; Huang, B. Research Progress in Computer-aided Diagnosis of Cancer Based on Deep Learning and Medical Images. J. Biomed. Eng. 2017, 2, 160–165.
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Int. Conf. Neural Inf. Process. Syst. 2012, 60, 1066.
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large Scale Image Recognition. Comput. Sci. 2014, 6, 1556.
- Szegedy, C.; Liu, Y. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9.
- He, K.W.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778.
- Salehi, S.; Erdogmus, D.; Gholipour, A. Auto-context Convolutional Neural Network(Auto-Net) for Brain Extraction in Magnetic Resonance Imaging. IEEE Trans. Med. Imaging 2017, 36, 2319–2330.
- Xue, J.; Hu, J.Y. Hypergraph Membrane System Based F2 Fully Convolutional Neural Network for Brain Tumor Segmentation. Appl. Soft Comput. J. 2020, 94, 106454.
- Isensee, F.; Wick, W.; Kickingereder, P.; Bendszus, M.; Maier, H.K. Brain Tumor Segmentation and Radio Mics Survival Prediction: Contribution to the BRATS 2017 Challenge. In Proceedings of the 3rd International Brain lesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Quebec City, QC, Canada, 10–14 September 2017; Springer: Berlin/Heidelberg, Germany, 2018; pp. 287–297.
- Saouli, R.; Akil, M.; Kachouri, R. Fully Automatic Brain Tumor Segmentation Using End-to-end Incremental Deep Neural Networks in MRI Images. Comput. Methods Programs Biomed. 2018, 166, 39–49.
- Zhang, J.; Lv, X. Ares U-Net: Attention Residual U-Net for Brain Tumor Segmentation. Symmetry 2020, 12, 721.
- Zhou, Z.X.; He, Z.S.; Shi, M.F.; Du, J.L.; Chen, D.D. 3D Dense Connectivity Network with Atrous Convolutional Feature Pyramid for Brain Tumor Segmentation in Magnetic Resonance Imaging of Human Head. Comput. Biol. Med. 2020, 121, 103766.
- Zhou, T.; Huo, B.Q.; Lu, H.L. Research on Residual Neural Network and Its Application in Medical Image Processing. Chin. J. Electron. 2020, 48, 1436–1447.
- Anand, V.K.; Grampurohit, S. Brain Tumor Segmentation and Survival Prediction Using Automatic Hard Mining in 3D CNN Architecture. arXiv 2021, arXiv:2101.01546v1.
- Havaei, M.; Davy, A.; Warde, F.D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.-M.; Larochelle, H. Brain Tumor Segmentation with Deep Neural Networks. Med. Image Anal. 2017, 35, 18–31.
- Lai, X.B.; Xu, M.S.; Xu, X.M. Multimodal MR Image Segmentation of Glioblastoma Based on Multi-class CNN. Chin. J. Electron. 2019, 47, 140–149.
- Hussain, S.; Anwar, S.M. Segmentation of Glioma Tumors in Brain Using Deep Convolutional Neural Network. Neuro Comput. 2018, 282, 248–261.
- Kamnitsas, K.; Ledig, C. Efficient Multi-Scale 3D CNN with Fully Connected CRF for Accurate Brain Lesion Segmentation. Med. Image Anal. 2017, 36, 61–78.
- Hu, K.; Deng, S.H. Brain Tumor Segmentation Using Multi-Cascaded Convolutional Neural Networks and Conditional Random Field. IEEE Access 2019, 7, 2615–2629.
- Zhao, X.; Wu, Y.; Song, G.; Li, Z.; Zhang, Y.; Fan, Y. A Deep Learning Model Integrating FCNNs and CRFs for Brain Tumor Segmentation. Med. Image Anal. 2017, 43, 98–111.
- Zhou, Z.X.; He, Z.S.; Jia, Y.Y. AFP-Net: A 3D Fully Convolutional Neural Network with Atrous-convolution Feature Pyramid for Brain Tumor Segmentation via MRI Images. Neuro Comput. 2020, 402, 03097.
- Liu, D.; Zhang, H.; Zhao, M.; Yu, X.; Yao, S.; Zhou, W. Brain Tumor Segmentation Based on Dilated Convolution Refine Networks. In Proceedings of the 16th IEEE International Conference on Software Engineering Research, Management and Application, Kunming, China, 13–15 June 2018; pp. 113–120.
- Badrinarayan, V.; Kendall, A. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495.
- Chen, S.C.; Ding, C.X.; Liu, M.F. Dual-force Convolutional Neural Networks for Accurate Brain Tumor Segmentation. Pattern Recognit. 2019, 88, 90–100.
- Zhou, Z.W.; Siddiquee, M.R. U-Net++: A Nested U-Net Architecture for Medical Image Segmentation. International Workshop on Deep Learning in Medical Image Analysis Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Cham, Switzerland; Berlin/Heidelberg, Germany, 2018; pp. 3–11.
- Alom, M.Z.; Hasan, M.; Yakopcic, C.; Taha, T.M.; Asari, K. Recurrent Residual Convolutional Neural Network Based on U-Net (R2U-Net) for Medical Image Segmentation. Comput. Vis. Pattern Recognit. 2018, 5, 06955.
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the International Conference on 3D Vision, Stanford, CA, USA, 25–28 October 2016; pp. 565–571.
- Hua, R.; Huo, Q.; Gao, Y.; Sui, H.; Zhang, B.; Sun, Y.; Mo, S.; Shi, F. Segmenting Brain Tumor Using Cascaded V-Nets in Multimodal MR Images. Front. Comput. Neurosci. 2020, 14, 9.
- Çiçek, Ö. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Cambridge, UK, 19–22 September 1999; pp. 424–432.
- He, C.E.; Xu, H. Research on Automatic Segmentation Algorithm for Multimodal MRI Brain Tumor Images. Acta Opt. Sin. 2020, 40, 0610001.
- Tseng, K.L.; Lin, Y.L.; Hsu, W.; Huang, C.Y. Joint Sequence Learning and Cross-modality Convolution for 3D Biomedical Segmentation. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 3739–3746.
- Li, H.C.; Li, A.; Wang, M.H. A Novel End-to-end Brain Tumor Segmentation Method Using Improved Fully Convolutional Networks. Comput. Biol. Med. 2019, 108, 150–160.
- Jia, H.Z.; Xia, Y. Learning High-Resolution and Efficient Non-Local Features for Brain Glioma Segmentation in MR Images; Medical Image Computing and Computer Assisted Intervention, MICCAI: Lima, Peru, 2020; pp. 480–490.




