Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Neuroimaging
脑肿瘤图像的精确分割是实现脑肿瘤准确诊断和有效治疗的重要一步。磁共振成像 (MRI) 可以生成没有组织损伤或颅骨伪影的脑图像,为临床医生在脑肿瘤和其他脑部疾病的研究中提供重要的判别信息。脑肿瘤MR图像的分割方法根据分割原理的不同主要分为三类:传统的分割方法、传统的基于机器学习的分割方法和基于深度学习的分割方法。
- image segmentation
- brain tumor
- magnetic resonance imaging
- multi-modality
1. 传统的脑肿瘤分割方法
1.1. 基于阈值的分割方法
Threshold-based segmentation is the simplest method. First, it is assumed that the pixels within a range belong to the same category [57]. Brain tumor images can be divided into target region and background region by setting an appropriate threshold. Different thresholds can also be set to divide the tumor into multiple regions. After continuous research and development, the accuracy of threshold segmentation has been greatly improved. Wang Y P et al. proposed an improved threshold segmentation algorithm. The method improves the noise sensitivity in threshold segmentation by using local information of pixel neighborhood [58]. Foladivanda et al. proposed an adaptive threshold segmentation method. The method can effectively overcome the problem of uneven gray, and enhance the contrast of images, and effectively improve the DSC and JS measure of MR image segmentation of the brain tumor [59].
The segmentation method based on threshold is relatively simple, and the quality of segmentation results almost entirely depends on the size of threshold, so the selection of threshold is very important. Moreover, the threshold segmentation method can only segment simple images, and it is difficult to deal with complex images.
1.2. Segmentation Methods Based on Region
Common region-based segmentation methods include watershed algorithm and region-growing algorithm.
Watershed algorithm is a segmentation method based on mathematical morphology. In this algorithm, the image to be processed is compared to the terrain in geography, and the elevation of terrain is represented by the gray value of the pixel. The local minimum and its adjacent area are called the ponding basin. It is assumed that there are water permeable holes at each local minimum. With the increase of infiltration water, the ponding basin will be gradually submerged. Blocking the flow of water from a stagnant basin to a nearby basin is called a dam. When the water level reaches the peak, the infiltration process ends. These dams are called watersheds. Kaleem et al. [60] proposed a watershed segmentation method guided by setting internal or external markers to calculate the morphological gradient of the input image and internal and external markers of the original image. Then they use watershed transform to obtain the segmentation results. Rajini N et al. [61] proposed a method combining threshold segmentation and watershed. First, the image was segmented by threshold method, and then the segmented image was segmented by watershed algorithm. The experiment proved that the segmentation results obtained by this method were more accurate than those obtained by one of the two methods alone, with the average TPR measure higher than 90%.
The segmentation algorithm based on watershed can obtain a complete closed curve and provide contour information for subsequent processing, whereas the watershed algorithm is influenced by noise and easy to over segment.
The region growing algorithm draws all the pixel points conforming to the criterion into the same region via formulating a criterion, so as to achieve pixel segmentation. This kind of segmentation method has the following characteristics: (1) Each pixel must be in a certain region, and the pixels in the region must be connected, and must meet certain similar conditions; (2) different regions are disjoint, and two different regions cannot have the same property. Qusay et al. [62] proposed an automatic seed region growth method, which can automatically set the initial value of seeds, avoid the defects of manual interaction, and improve the efficiency of image segmentation.
The region-based segmentation method has the characteristics of simple calculation and high accuracy, which can extract better regional features and is more suitable for segmentation of small targets. However, it is sensitive to noise and easy to make holes in the extracted region.
1.3. Segmentation Methods Based on Fuzzy Theory
The segmentation methods based on fuzzy theory have also been highly valued. In brain tumor MR image segmentation, the most widely used Fuzzy theory algorithm is Fuzzy C-means clustering (FCM) [63]. Muneer K et al. [64] obtained the K-FCM method through the combination of FCM algorithm and K-means algorithm. The experiment proved that, compared with FCM, K-FCM showed higher accuracy in brain tumor MR image segmentation and could reduce the computational complexity. Guo Y et al. [65] proposed a Neutrosophic C-Means (NCM) algorithm based on fuzzy C-means and neutral set framework. The algorithm introduced distance constraint into the objective function to solve the problem of insufficient prior knowledge and achieved satisfactory segmentation results. On the basis of Super-pixel fuzzy clustering and the lattice Boltzmann method, Asieh et al. [66] proposed a level set method that can automatically segment brain tumors, which has strong robustness to image intensity and noise.
The segmentation method based on fuzzy theory can effectively solve the problem of incomplete image information, imprecision, and so on. It has strong compatibility and can be used in combination with other methods, but it is difficult to deal with large-scale data due to its large amount of computation and high time complexity.
1.4. Segmentation Methods Based on Edge Detection
The segmentation principle based on edge detection and target contour achieves segmentation by obtaining the edge of the target region and then obtaining the contour of the target region. Common detection operators for edge detection include Roberts operator, Sobel operator, Canny operator and Prewitt operator [67]. Jayanthi et al. [68] integrated FCM into the active contour model. The initial contour of the model is automatically selected by FCM, which reduces the human–computer interaction. Moreover, the problem of the unclear edge contour and uneven intensity in MR images was improved. The average DSC measure of segmentation by this method reached 81%.
Compared with other traditional segmentation methods, the segmentation method based on edge detection pays attention to the edge information of the image and links the edges into contours, and the anti-noise performance is stronger. But the anti-noise performance is negatively correlated with accuracy, that is, the better the anti-noise performance, the lower the accuracy. On the contrary, improved accuracy will reduce the anti-noise performance.
2. Segmentation Methods of Brain Tumor MR Images Based on Traditional Machine Learning
Brain tumor segmentation methods based on traditional machine learning use predefined features to train the classification model. Generally, they are divided into two levels: organizational level and pixel level. At the organizational level, the classifier needs to determine which kind of organizational structure each feature belongs to, and at the pixel level the classifier needs to determine which category each pixel belongs to. Traditional Machine Learning algorithms mainly include K-Nearest Neighbors (KNN) [69], Support Vector Machine (SVM) [70], Random Forest (RF) [71], Dictionary Learning (DL) [72], etc.
Havaei et al. [69] regarded each brain as a separate database and used the KNN algorithm for segmentation. They obtained very accurate results, and the segmentation time of each brain image is only one minute, which improves the efficiency of segmentation. Llner F et al. [70] used SVM to segment brain tumors, taking into account the changing characteristics of signal intensity and other features of brain tumor MR images. The TPR measure of this method for LGG reached 83%, and the accuracy measure for HGG reached 91%. Sher et al. [73] first segmented the image by the Otsu method and K-means clustering, then extracted the features by discrete wavelet transformation, and finally reduced the feature dimension by the PCA algorithm to obtain the best features for SVM classification. The experimental results show that the sensitivity and specificity of the scheme can reach more than 90%. Vaishnavee et al. [74] used a proximal support vector machine (PSVM). The method uses equation constraints to solve the primary linear equations, which simplifies the original problem of solving convex quadratic programming. The experiment shows that PSVM is more accurate than SVM in MR image segmentation of brain tumor. Wu et al. [75] proposed a method to first segment the image into super-voxels, then segment the tumor using MRF, estimate the likelihood function at the same time, and extract the features using a multistage wavelet filter. Nabizadeh et al. [76] proposed an automatic segmentation algorithm based on texture and contour. Firstly, the initial points were determined and the machine learning classifier was trained by the initial points. Mahmood et al. [71] proposed an automatic segmentation algorithm based on RF. This algorithm uses several important features such as image intensity, gradient and entropy to generate multiple classifiers, and classifies pixels in multispectral brain MR images by combining the results to obtain segmentation results. Selvathi et al. [77] increased the weight of the wrongly classified samples and decreased the weight of the correctly classified samples in the training process. Then the classifier gives new weights to the samples to ensure that the weights of all decision trees are positively correlated with their classification ability. Finally, the input of the improved RF consists of two parts: the image intensity feature and the original image feature extracted by curve and wavelet transformation. Experimental results show that the accuracy of the improved RF scheme is 3% higher than that of the original RF algorithm. Reza et al. [78] studied the correlation of image minimization features from the perspective of image features, effectively selected features, and finally classified features in multimodal MR images through RF. Compared with the RF algorithm alone, the proposed method can improve the DSC, PPV and TPR measure simultaneously. Meier et al. [79] trained a specific random forest classifier by semi-supervised learning. It takes image segmentation as a classification task and effectively combines the preoperative and postoperative MR image information to improve the postoperative brain tumor segmentation. The PPV and ME measure obtained by this method were 93% and 2.4%, respectively. Dictionary learning is a kind of learning method for simulating dictionary lookup. The dictionary itself is set as dictionary matrix, and the method used is sparse matrix. The process of dictionary lookup is obtained by multiplying the sparse matrix and dictionary matrix, and then the dictionary matrix and sparse matrix are optimized to minimize the error between the value searched and the original data. Chen et al. [72] transformed the super-pixel feature into a high-dimensional feature space. According to the different error values of different regions when the dictionary was modeling brain tumors, the segmentation of brain tumor MR images was realized and the segmentation accuracy was improved. Li [80] proposed a multi dictionary fuzzy learning algorithm based on dictionary learning. This algorithm effectively combines dictionary learning with fuzzy algorithm, and fully considers the differences between the target region and the background, as well as the consistency within the target region. This method can describe the gray and texture information of different regions of the image, and segment the image quickly and accurately.
The traditional machine learning algorithm is better than many traditional segmentation algorithms in algorithmic performance, but there are many shortcomings when it is used in brain tumor MR image segmentation. For example, the KNN algorithm is simple to implement, and the prediction accuracy of the brain tumor region is relatively high, but the calculation is relatively large [69]. The support vector machine has strong theory, and the final result is determined by several support vectors. The calculation is relatively simple and the generalization ability is strong, but it has higher requirements concerning the selection of parameters and kernel function [70]. Random forest can solve the problem of over-segmentation well, process multiple types of data, and has good anti-noise performance. It can parallel operation and shorten the operation time, but it has a poor effect on low-dimensional tumor data processing [71]. The algorithm based on dictionary learning is similar to the idea of dimensionality reduction, both of which reduce the computing complexity and speed up the computing speed, but also have higher requirements for tumor data [72].
3. Segmentation Methods of Brain Tumor MR Images Based on Deep Learning
3.1. Segmentation Methods of Brain Tumor MR Images Based on CNN
Convolutional neural network belongs to the category of neural network, and its weight sharing mechanism greatly reduces the model complexity. Convolutional neural network (the network diagram is shown in Figure 1a) can directly take the image as the input, automatically extract the features, and has a high degree of invariance to the image translation, scaling and other changes. In recent years, a series of Network models based on convolutional neural Network [81], such as Network in Network [82], VGG [83], Google-Net [84], Res-Net [85], etc., have been widely used in medical image segmentation. Among them, the VGG network has a strong ability to extract features and can guarantee the convergence in the case of fewer training times. However, as the deepening of the network will cause gradient explosion and gradient disappearance, the optimization effect will start to deteriorate when the network depth exceeds a certain range.
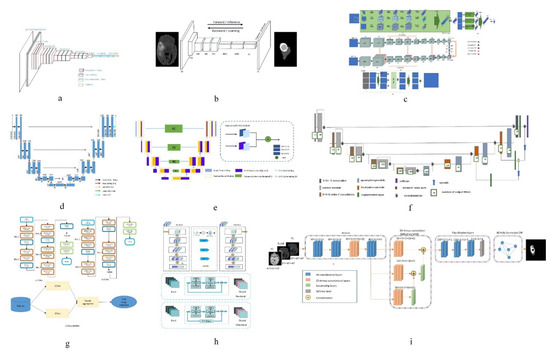
Figure 1. Network structure diagrams of some classical methods and improved methods. (a) is the classic CNN network model; (b) is the classic FCN network model; (c) is the structure diagram of a convolutional neural network technology based on automatic context (Auto-Nets) proposed by Salehi et al. [6]; (d) is the classic Encoder-Decoder network model; (e) is the structure diagram of a fully convolutional neural network with feature reuse module and feature integration module (f2fcn) proposed by Xue et al. [90]; (f) is the structure diagram of a robust neural network algorithm based on u-net proposed by isensee et al. [31]; (g) is the structure diagram of a sequential CNN architecture proposed by saouli et al. [89]; (h) is the structure diagram of attention residual U-net proposed by Zhang et al. [38]; (i) is a structural diagram of 3D dense connection combined with feature pyramid proposed by Zhou et al. [12].
In order to solve the problem of network degradation, He et al. [85] proposed deep Residual Network (ResNet), which achieved good results in the segmentation task [86]; Anand et al. [50] combined the 3D convolutional neural network with dense connection, pre-trained the model, and then initialized the model with the weight obtained. This method improved the DSC measure in the segmentation task of brain tumor MR images. Havaei et al. [18] constructed a cascaded dual path CNN, which took the output characteristic graph of CNN in the first stage as the additional input of CNN in the second stage. This method can effectively obtain rich background information and get better segmentation results. Lai et al. [87] reduced the tail of the original image by 98% firstly, corrected the bias field by using n4itk, then pre-segmented it by multi classification CNN, and finally obtained the final segmentation result by median filtering. The algorithm improves the DSC and PPV of segmentation significantly. Salehi et al. [6] proposed a convolutional neural network technology based on automatic context (Auto-Nets) to indirectly learn 3D image information by means of 2D convolution. This method uses 2D convolution in axial, coronal and sagittal MR images respectively to avoid complex 3D convolution operations in segmentation (The network diagram is shown in Figure 1c). Hussain et al. [88] established a correlation architecture composed of a parallel CNN layer and a linear CNN layer by adding an induction structure. This structure has achieved good results in brain tumor MR image segmentation, especially in enhancing the DSC measure to 90%. Kamnitsas et al. [24] trained 3D brain tumor images and then carried out conditional random field post-processing to obtain smoother results. Saouli et al. [89] designed a sequential CNN architecture and proposed that an end-to-end incremental network can simultaneously develop and train CNN models (the network diagram is shown in Figure 1g). The average DSC measure obtained by this method is 88%. Hu K et al. [22] proposed a more hierarchical convolution based Neural Network (Multi-Cascaded Convolutional Neural Network, MCCNN) and fully connected conditional random fields (CRFs), combined with the brain tumor segmentation method, Firstly, the brain tumor is roughly segmented by multi classification convolution neural network, and then fine segmented by fully connected random field according to the rough segmentation results, so as to achieve the effect of batch segmentation and improve the accuracy. The segmentation algorithm based on CNN can automatically extract features and process high-dimensional data, but it is easy to lose information in the process of pooling, and its interpretability is poor.
3.2. Segmentation Methods of Brain Tumor MR Images Based on FCN
Compared with pixel-level classification, image-level classification and regression tasks are more suitable for using the CNN structure, because they both expect to obtain a probable value for image classification. For semantic segmentation of images, FCN works better. FCN has no requirement on the size of the input image, and there will be an up sampling process at the last convolution layer. This process can get the same result as the input image size, predicting each pixel while retaining the spatial information in the input image, so as to achieve the pixel classification. In simple terms, FCN is a method to classify and segment images at the pixel level. Therefore, the semantic segmentation model based on FCN is more in line with the requirements of medical image segmentation. Zhao et al. [20] proposed a combination of FCN with CRF for brain tumor segmentation. The method trains two-dimensional slices in axial, coronal and sagittal directions respectively, and then uses fusion strategy to combine segmented brain tumor images. Compared with the traditional segmentation methods, the segmentation speed is faster and the efficiency is higher. Xue et al. [90] proposed a fully convolutional neural network with feature reuse module and feature integration module (f2fcn). It reuses the features of different layers, and uses the feature integration module to eliminate the possible noise and enhance the fusion between different layers (the network diagram is shown in Figure 1e). The DSC and PPV obtained by this method are high. Zhou et al. [91] proposed a 3D atomic convolution feature pyramid to enhance the discrimination ability of the model, which is used to segment tumors of different sizes. Then, an improvement is made on the original basis [12], a 3D dense connection architecture is proposed, and a new feature pyramid module is designed by using 3D convolution (the network diagram is shown in Figure 1i). This module is used to fuse multi-scale context to improve the accuracy of segmentation. Liu et al. [26] proposed a Dilated Convolution optimization structure (DCR) based on Resnet-50, which can effectively extract local and global features, and this method can improve the segmentation PPV measure to 92%. The segmentation algorithm based on FCN can predict the category of each pixel, transform the image classification level to the semantic level, retain the position information in the original image, and obtain a result with the same size as the input image. However, the algorithm has low computational efficiency, takes up a lot of memory space, and the receptive field is relatively small.
3.3. Segmentation Methods of Brain Tumor MR Images Based on Encoder-Decoder Structure
The encoder-decoder structure is generally composed of an encoder and a decoder. The encoder trains and learns the input image through a neural network to obtain its characteristic map. The function of the decoder is to mark the category of each pixel after the encoder provides the feature map, so as to achieve the segmentation effect. In the segmentation tasks based on encoder-decoder structure, the structure of encoders is generally similar, mostly derived from the network structure of classification tasks, such as VGG, etc. The purpose of doing this is to obtain the weight parameters of network training through the training of a large database. Therefore, the difference of the decoder reflects the difference of the whole network to a large extent, and is also the key factor affecting the segmentation effect.
Badrinarayanan 等人。[ 92] 提出了 SegNet 模型。与其他模型相比,该模型具有更深的层次,在像素的语义分割方面具有更好的性能。模型的编码器部分由一个13层的vgg-16网络组成,可以记住编码阶段最大像素的位置信息。在解码器中,对低分辨率输入特征进行上采样以获得分割结果。基于 FCN 的 U-Net 模型是一种广泛使用的脑肿瘤分割模型,其中网络结构也由编码器和解码器组成,U-Net 网络跳转连接会对路径进行编码,用于得到把图中的特征特征放到解码路径对应的位置,以便得到编码阶段下直接采样的特征进入解码阶段,从而学习更详细的特征。陈等人。[[93 ]提出了一种多级深度网络,可以通过在Multi-Level Deep Medical (MLDM)和U-Net上添加辅助分类器来获取图像多级信息,从而实现图像分割。DSC、PPV和TPR的结果分别为83%、73%和85%。为了减少编码器和解码器网络的特征映射之间的语义差距,Zhou等人。[ 94 ]提出了多种嵌套密集连接方法来连接编码器和解码器网络。阿洛姆等人。[ 95 ]提出了基于U-Net的递归神经网络和递归残差卷积神经网络。实验结果表明,两种网络分割结合U-Net的性能优于单独使用U-Net。张等人。[38 ]引入了注意机制和残余网络进入传统的U Net网络和提出一种关注残余U形网(网络图被示出在 图1 h)中,其中改进的脑肿瘤MR图像的分割性能。米勒塔里等人。[ 96 ] 在 3D U-Net 模型的基础上提出了 V-Net 模型,它通过使用 3D 卷积检查扩展了原始 U-Net 模型。华等人。[ 37 ]级联V-Net,采用先将整个肿瘤分割成肿瘤子区域的方法;分割的准确率高于直接 V-Net 分割。西切克等人。[ 97] 提出了一个 3D U-Net 模型来学习稀疏注释体积图像的特征。在 3D U-Net 的基础上,Heet 等人。[ 98 ]添加了一个混合扩张卷积(HDC)模块来增加神经元的感觉场,克服多尺度特征提取需要深度神经网络的限制。使用浅层神经网络可以减少模型参数的数量,降低计算复杂度。Tsenget 等人。[ 25]提出了一种具有深度的跨模态卷积编码器/解码器结构,结合不同模态的MR图像数据,同时采用加权和多阶段训练的方法来解决数据不平衡的问题;与传统的U-Net结构相比,改进了DSC、TPR和PPV测量方法。伊森西等人。[ 31 ]改善了U形Net网络模型并设计了一种稳健的神经网络算法,它防止通过扩大数据量过拟合(网络图被示出在 图1中F)。该算法将 TPR 测量提高到 91%;海春等。[ 28] 巧妙地将改进的全卷积神经网络结构应用于U-Net模型,提出了一种新颖的端到端脑肿瘤分割方法。该方法在编码路径和解码路径之间设计了一种上跳连接结构,以增强信息流。贾等人。[ 99]构建了基于并行多尺度融合(PMF)模块的HNF网络,并提出了用于多参数MR成像的三维高分辨率非局部特征网络(HNF-NET),该网络可以产生强大的高分辨率特征表示和聚合多尺度上下文信息。引入了期望最大化注意力(EMA)模块以提取更多相关特征并减少冗余特征。整个肿瘤的DSC和HD分别为91.1%和4.13%。基于encoder-decoder的分割算法可以结合高分辨率和低分辨率信息,可以从多个尺度识别特征,但是编码过程和解码过程之间只有很短的联系,两者之间的联系是显然不够。
This entry is adapted from the peer-reviewed paper 10.3390/healthcare9081051
This entry is offline, you can click here to edit this entry!
