Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Arumugam Radhakrishnan Jayakumar | + 2654 word(s) | 2654 | 2021-09-03 08:11:37 | | | |
| 2 | Rita Xu | Meta information modification | 2654 | 2021-09-13 04:54:18 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Jayakumar, A. Multi-Organ Histopathological Changes. Encyclopedia. Available online: https://encyclopedia.pub/entry/14122 (accessed on 13 January 2026).
Jayakumar A. Multi-Organ Histopathological Changes. Encyclopedia. Available at: https://encyclopedia.pub/entry/14122. Accessed January 13, 2026.
Jayakumar, Arumugam. "Multi-Organ Histopathological Changes" Encyclopedia, https://encyclopedia.pub/entry/14122 (accessed January 13, 2026).
Jayakumar, A. (2021, September 13). Multi-Organ Histopathological Changes. In Encyclopedia. https://encyclopedia.pub/entry/14122
Jayakumar, Arumugam. "Multi-Organ Histopathological Changes." Encyclopedia. Web. 13 September, 2021.
Copy Citation
Infection with SARS-CoV-2, the virus responsible for the global COVID-19 pandemic, causes a respiratory illness that can severely impact other organ systems and is possibly precipitated by cytokine storm, septic shock, thrombosis, and oxidative stress. SARS-CoV-2 infected individuals may be asymptomatic or may experience mild, moderate, or severe symptoms with or without pneumonia.
COVID-19
SARS-CoV-2
mice
multiorgan histopathology
mouse hepatitis virus-1
1. Introduction
Infection with SARS-CoV-2, the virus responsible for the global COVID-19 pandemic, primarily causes a respiratory illness. Approximately 193 million cases have been reported worldwide, resulting in more than 4 million deaths to date. COVID-19 is likely precipitated by cytokine storm, septic shock, thrombosis, and oxidative stress [1]. Infected individuals develop a wide range of symptoms ranging from mild to severe illness. Symptoms can appear 2–14 days after exposure to the virus. These include, fever, chills, loss of taste or smell, sore throat and cough, shortness of breath or difficulty in breathing, congestion and runny nose, fatigue, muscle and body aches, severe headache, nausea or vomiting, and diarrhea [2][3].
Most individuals (81%) develop mild to moderate symptoms of COVID-19, which can extend to pneumonia [4][5][6][7][8][9][10][11]. However, 14% of patients develop severe symptoms that include dyspnea and hypoxia, with more than 50% showing lung involvement [12][13]. Additionally, 5% of patients suffer critical symptoms such as respiratory failure and shock. At least one third of individuals infected with SARS-CoV-2 virus do not develop noticeable symptoms at any point in time [14]. Asymptomatic carriers tend to not get tested, thus spreading the virus to others and leading to an increase in infections. Individuals may develop symptoms later (pre-symptomatic) or have very mild symptoms [14]. Recently, multiple SARS-CoV-2 variants were reported to be circulating globally. These include a UK variant (B.1.1.7, also known as VOC-202012/01), South African variants (B.1.351, 501Y.V2), and Brazilian variants of SARS-CoV-2 (P.1, B.1.1.28.1, and B.1.617.2). These variants have been classified by the CDC as variants of concern [15] due to their high rate of transmissibility, although their epidemiology and impact on humans are unclear. More recently, the additional SARS-CoV-2 variants B.1.427, B.1.429, B.1.525, P.3, B.1.617.1, B.1.620, B.1.621, and C.37 (variants of interest), as well as B.1.617.3, B.1.214.2, A.23.1+E484K, A.27, A.28, C.16, B.1.351 + P384L, B.1.351 + E516Q, B.1.1.7 + L452R, B.1.1.7 + S494P, C.36 + L452R, AT.1, B.1.526, B.1.526.1, B.1.526.2, B.1.1.318, P.2, B.1.1.519, AV.1, P.1 + P681H, B.1.671.2 + K417N (variants under monitoring), and B.1.427/B.1.429 and B.1.616 (de-escalated variants) have been reported.
Over the past 1000 years, coronaviruses have continually evolved [16]. The earliest identification of coronaviruses was in animals. It first isolated as an infectious bronchitis virus (IBV) in chickens in 1947 [17], followed by the isolation of mouse hepatitis virus (MHV) in mice in 1949 [18]. In 1946, pigs in the United States were identified as carriers of a transmissible gastroenteritis virus (TGEV) [18]. In the 1960s, human coronaviruses were isolated from respiratory tract infections [19]. B814 and 229E were the first two viruses isolated in humans [20][21]. Subsequently, tissue cultures were used to isolate additional coronavirus strains (OC16 and OC43) from humans [22][23]. To date, coronaviruses have been identified in numerous other species, including calves, dogs, cats, bats, sparrows, rabbits, and turkeys [24]. Early phylogenetic studies on SARS-CoV-2 genomic sequences showed that it clustered closely with sequences originating from SARS-like viruses from bats within lineage B of the betacoronavirus genus. Lineage A groups are prototypical coronaviruses such as MHV and the human coronaviruses HCoV-HKU1 and HCoV-OC43 (Figure 1A,B) ([25][26][27][28][29][30][31][32][33][34][35][36][37][38][39][40] tree.bio.ed.ac.uk/software/figtree/ accessed on 7 July 2021) (Software (ed.ac.uk accessed on 7 July 2021)), while the other highly pathogenic coronavirus, MERS-CoV, is found within lineage C, along with the related camel-derived MERS-CoV ([41][42] and references therein). The S protein amino acid sequences from four representative beta coronaviruses (HCoV-HKU1, MHV, SARS-CoV, and MERS-CoV) were aligned and the solved S protein structures were compared to determine their amino acid identity and the overall structural organization similarities among these proteins. The findings showed an average of approximately 30% identity among the four viral S proteins at the amino acid level, with the exception of HCoV-HKU1 and MHV, which share an amino acid identity of 59% at the S protein ([41][42] and references therein). While there are dissimilarities between the amino acid sequences, the structure of the four betacoronavirus S proteins were found to have similar folding patterns.
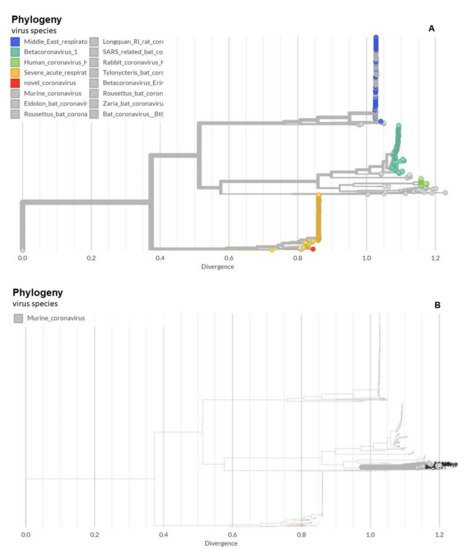
Figure 1. Phylogeny of coronaviruses including the novel coronavirus SARS-CoV-2. (A) Phylogeny of coronaviruses including the novel coronavirus SARS-CoV-2.; (B) Phylogeny of murine coronaviruses including the murine hepatitis virus.
MHV-1 infection of A/J mice produces clinical SARS-like disease with high mortality [43][44][45]. These mice develop severe pulmonary disease at day 6 post-MHV-1 infection and 60% mortality from 7 to 12 days post-infection. On day 2, patchy interstitial alveolar thickening and fluid accumulation in alveolar spaces (pulmonary edema) are prominent. At death, the lungs show severe interstitial pneumonitis with large areas of complete consolidation. The interstitial inflammatory reaction includes the presence of hyaline membranes, fibrin deposition, and significant lymphocyte and macrophage infiltrates. Infiltrating cells in MHV-1-infected lung tissue from A/J mice are predominantly macrophages and neutrophils on days 2–6 post-MHV-1 infection. T cells (CD3 positive) also increase in the infiltrates, especially by day 6 post-infection. Examinations of the livers of MHV-1-infected A/J mice show normal histology at day 6, but on days 7 or 8 just prior to death, evidence of severe hepatic congestion has been observed, as in humans [46]. Thus, A/J mice are highly susceptible to MHV-1-induced pulmonary disease when the virus is delivered intranasally.
2. Animal Survival Post-MHV-1 Infection
MHV-1 inoculated female mice displayed signs of sickness (drowsiness and lack of movement) at 2 days post-exposure. About 43.75% of mice (7/16) showed signs of illness on day 2 post-MHV-1 inoculation, and the number of mice exhibiting clinical signs increased gradually (56.25%, 62.5%, and 75.0% on days 3, 4, and 5, respectively) (Figure 2A). Of note, the remaining 25% of mice did not show any clinical signs (observed for up to 91 days). Further, exposure of mice to DMEM alone had no effect on animal survival.
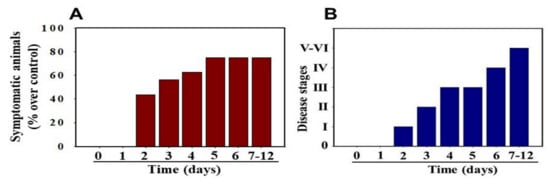
Figure 2. MHV-1 inoculated mice displayed signs of sickness. (A) About 40% of mice showed signs of sickness on day 2 post-MHV-1 inoculation, and the number of mice exhibiting clinical signs increased gradually (up to 75%). The remaining 25% of mice did not show any clinical signs (observed for up to 91 days). (B) MHV-1 inoculated mice showed clinical signs (from stages I to VI) from days 2 to 12 and beyond. Mild to moderate clinical signs (stages I–III) were observed from days 2 to 4. The MHV-1 infected mice showed severe sickness (stages IV–VI) from day 6 onward. Further, 60% of animals died from days 7 to 12 (n = 16).
Stage I symptoms (drowsy and lack of movement) on day 2 and stage II symptoms (slightly ruffled fur and altered hind limb posture) on day 3 were observed post-MHV-1 inoculation. Stage III symptoms (ruffled fur and mildly labored breathing) were detected on days 4 and 5. Stage IV (ruffled fur, inactivity, moderately labored breathing, and tremor) was observed on day 6, and stages V and VI (ruffled fur, obvious labored breathing and lethargy, as well as a moribund state and death) were noted from days 7 to 12 (Figure 2B). Figure 2A,B involves the same animals. For example, in Figure 2A, approximately 40% of animals showed clinical signs of infection and all of them were in stage I. On day 3, about 55–60% animals showed clinical signs of infection that were of stage II. The additional mice (10–20%) that exhibited clinical signs of infection at later stages showed a severity similar to that of mice that exhibited clinical signs of infection at early stages. Of note, we observed mild diarrhea (several loose stools in one day) in the early stages of illness (from 2 to 4 days). However, the diarrhea ended from days 5 to 12 and occurred only in a few animals (5/16 mice), as observed in humans in association with COVID-19 [47][48]. Further, MHV-1 inoculated mice displayed severe venous thrombosis (4/16 mice at days 5–7), a characteristic feature of COVID-19 (Figure 3) [38][39][40][41][42][49][50][51][52][53]. Milder forms of thrombosis were also observed in MHV-1 inoculated mice at days 5–7 (7/16). The natural history of the disease was observed in mice that survived after day 12 for up to 91 days.
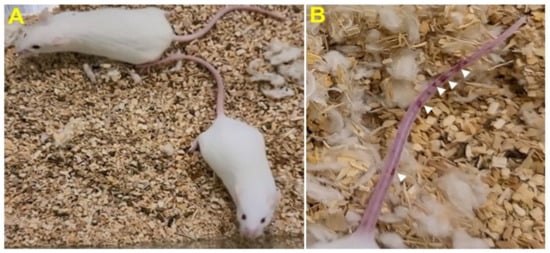
Figure 3. MHV-1 inoculated mice displayed severe disease. (A) Normal mouse. (B) A representative mouse from the MHV-1 inoculated group showing venous thrombosis consistent with symptoms in patients with COVID-19 (n = 11).
3. Weight Loss Post-MHV-1 Infection
MHV-1 inoculated mice were monitored for weight loss from day 2 onwards. MHV-1 administered mice showed approximately 20% weight loss on day 3, around 25–30% weight loss on day 4 and 5, and 35–40% weight loss from days 7 to 12 (Figure 4). Of note, the weight loss occurred rapidly on days 6–7 in animals that showed severe clinical signs (from about 25% to 40%). We were unable to control this since it was not obvious whether any particular animal would lose that much weight in a short period of time, and as such we could not provide glucose/KCl (i.e., fluid support). The reason for such rapid weight loss is unknown. It is possible that rapid body fluid loss or a lack of fluid intake and loss of muscle mass and fat during the end stages may have occurred, as has frequently been observed in humans in association with SARS-CoV-2 infection. While our elaborated survival/characterization study shows the exact nature of the viral infection, and our initial characterization study will assist researchers in better understanding the severity of disease progression and in designing mechanistic studies, we will include the humane endpoint criteria in future investigations based on our current observations.
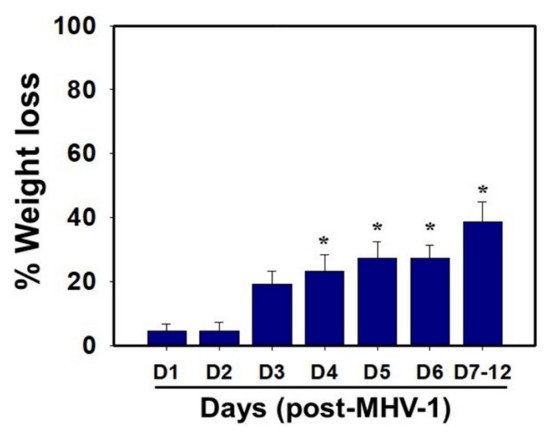
Figure 4. Body weight losses in MHV-1 infected mice. MHV-1 inoculated mice lost 20–40% of body weight over days 3–8, corresponding well with the severity of the disease (n = 16). * p < 0.05 versus control.
We examined the status of liver enzymes. Increased liver enzymes (ALT and AST) were identified in MHV-1-infected mice, as observed in patients with drug or chemical-induced acute liver failure or in association with COVID-19. AST levels increased manifold in MHV-1 inoculated mice as compared to un-inoculated mice (3459.2 ± 684.1 units/L in MHV-1 infected mice as compared to 96.8 ± 14.2 units/L in the control group, 34.7-fold increase over control). There was also an increase in ALT levels (3068.5 ± 861.3 units/L in MHV-1 infected mice as compared to 31.5 ± 11.6 in the control group, 96.4-fold increase over control). ALP and bilirubin levels were also increased in infected mice (986.3 ± 178.4 units/L of ALP in MHV-1 infected mice as compared to 589.1 ± 108.7 in the control group, 67% increase over control; and 0.86 ± 0.2 mg/L of bilirubin, as compared to 0.075 ± 0.02 mg/L in uninfected mice, 10.4-fold increase over control) (Table 1). Furthermore, the exposure of mice to DMEM had no effect on alterations in liver enzymes (i.e., levels were identical to those of healthy controls). These findings strongly suggest that MHV-1 leads to severe liver injury; similar observations have been made in patients with SARS-CoV-2 infection [46][54][55][56][57][58].
Table 1. Liver enzymes of mice exposed to MHV-1.
| Uninfected Mice | MHV-1 Infected Mice | |
|---|---|---|
| AST (Units/L) ALT (Units/L) ALP (Units/L) |
96.8 ± 14.2 31.5 ± 11.6 589.1 ± 108.7 |
3459.2 ± 684.1 * 3068.5 ± 861.3 * 986.3 ± 158.4 * |
| Bilirubin (mg/L) | 0.075 ± 0.02 | 0.86 ± 0.2 * |
4. Histopathological Changes Post-MHV-1 Infection
Lung: MHV-1-infected mice at day 7 showed inflammation (i.e., granular degeneration of cells, and migration of leukocytes into the lungs), along with proteinaceous debris filling of the alveolar spaces with fibrillar to granular eosinophilic protein strands caused by the progressive breakdown of the capillary wall and epithelial integrity, permitting leakage of protein rich edematous fluid into the alveoli, and the presence of hemosiderin-laden macrophages (indicating pulmonary congestion with dilated capillaries and leakage of blood into alveolar spaces). Furthermore, peribronchiolar interstitial infiltration, bronchiole epithelial cell necrosis, necrotic cell debris within alveolar lumens, alveolar exudation, hyaline membrane formation, alveolar hemorrhage with red blood cells within the alveolar space, and interstitial edema are all characteristic features of infected lungs in humans with SARS-CoV-2 infection [59] (Figure 5).
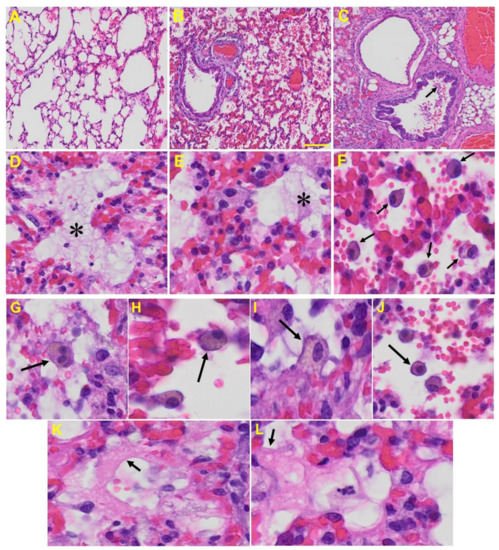
Figure 5. Lung from MHV-1 exposed mice. (A) Normal mouse. (B) MHV-1-infected mouse lung at day 7. (C) The MHV-1 infected mouse lung shows inflammation, granular degeneration of cells, and migration of leukocytes into the lungs (arrow). (D,E) Proteinaceous debris filling of the alveolar spaces with fibrillar to granular eosinophilic protein strands caused by progressive breakdown of the capillary wall and epithelial integrity, permitting the leakage of protein-rich edema fluid into the alveoli (asterisk) (commonly seen in ARDS). (F–J) Presence of hemosiderin-laden macrophages, indicating pulmonary congestion with dilated capillaries and leakage of blood into alveolar spaces. Further, peribronchiolar interstitial infiltration, bronchiole epithelial cell necrosis, necrotic cell debris within alveolar lumens, alveolar exudation, infiltration, hyaline membrane formation (K,L), and alveolar hemorrhage with red blood cells within the alveolar space and interstitial edema were also observed in these mice (H&E original magnification 400× Figure 5A–C; scale bar image divided by actual scale bar length).
Liver: Liver from MHV-1 exposed mice showed hepatocyte degeneration, severe periportal hepatocellular necrosis with pyknotic nuclei, severe hepatic congestion, ballooned hepatocytes, vacuolation, and the presence of piecemeal necrosis, as well as hemorrhagic changes. Ground glass hepatocytes showed voluminous, abundant, granular cytoplasm, peripheral cytoplasmic clearing and central nuclei, and apoptotic (acidophil) bodies, as well as absent hepatocytes replaced by abundant inflammatory cells. Condensation and dark staining of the cytoplasm, an absence of the nucleus, fatty changes, binucleated hepatocytes, and activated Kupffer cells were also observed in MHV-1 exposed mice livers (Figure 6).
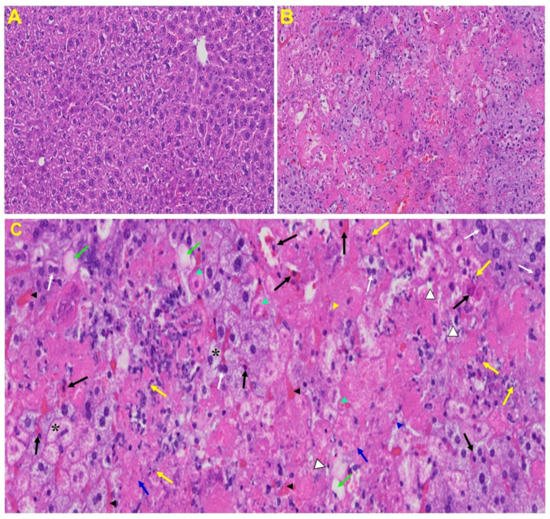
Figure 6. Liver from MHV-1 exposed mice. (A) Normal mouse. (B) Infected mouse liver at day 7. (C) The MHV-1 infected mouse liver at day 7 shows hepatocyte degeneration (yellow arrows), severe periportal hepatocellular necrosis with pyknotic nuclei (black arrows), severe hepatic congestion (black arrowheads), ballooned hepatocytes (asterisks), vacuolation (green arrows), and the presence of piecemeal necrosis (blue arrowhead), as well as hemorrhagic changes. Ground glass hepatocytes show voluminous, abundant, granular cytoplasm, peripheral cytoplasmic clearing and central nuclei, and apoptotic (acidophil) bodies. Several hepatocytes are absent and have been replaced by abundant inflammatory cells (yellow arrowhead). Note the condensation and dark staining of the cytoplasm and the absence of the nucleus (green arrowheads), fatty changes (blue arrows), binucleated hepatocytes (white arrows), and activated Kupffer cells (white arrowheads), indistinctive of severe liver failure (H&E original magnification of A&B is 400× as noted in Figure 5B).
Brain: Upon examination of the MHV-1 infected mice brain (Figure 7B,C), we observed congested blood vessels, perivascular cavitation (suggestive of edema), pericellular halos, vacuolation of neuropils, darkly stained nuclei and pyknotic nuclei amid associated vacuolation of the neuropil, and acute eosinophilic necrosis. The brain hippocampus of MHV-1 infected mice showed necrotic neurons with fragmented nuclei and vacuolation (Figure 7).
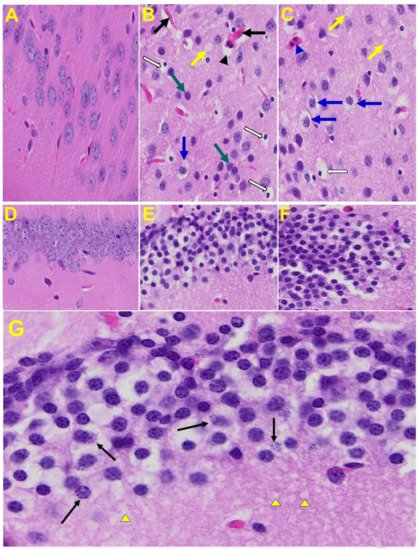
Figure 7. MHV-1 infected mouse brain. (A) Normal mouse. (B,C) The infected mouse brain cortex at day 7 shows congested blood vessels (black arrows), perivascular cavitation (black arrowhead) suggestive of edema, pericellular halos (blue arrows), vacuolation of neuropils (yellow arrow), darkly stained nuclei (curved arrows green arrows), pyknotic nuclei amid associated vacuolation of the neuropil (white arrows), and acute eosinophilic necrosis (blue arrowhead). (D) Normal mice. (E,F) The brain hippocampus of an MHV-1 infected mouse (enlarged image, (G)) shows a necrotic neuron with fragmented nucleus (arrow) and vacuolation (arrowhead) (H&E original magnification of A–F is 400× as noted in Figure 5B).
Heart: The heart of MHV-1 infected mice showed severe interstitial edema, vascular congestion and dilation, and red blood cells infiltrating between degenerative myocardial fibers (Figure 8).
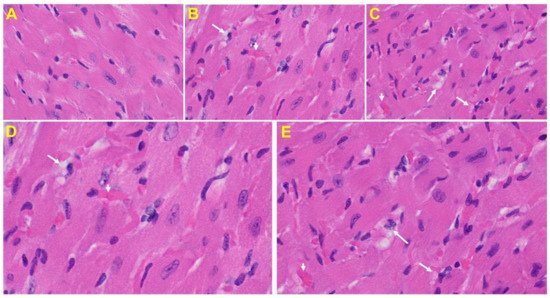
Figure 8. An MHV-1 infected mouse heart. (A) Normal mouse heart. (B,C) MHV-1 infected mouse heart showing severe interstitial edema (long arrows), vascular congestion and dilation (short arrows), and red blood cell extravasation into the interstitium (enlarged images of an MHV-1 infected mouse heart (D,E)) (H&E original magnification of A–C is 400× in A–C as noted in Figure 5B).
Kidney: Tubular epithelial cell degenerative changes, peritubular vessel congestion, proximal and distal tubular necrosis, hemorrhage in interstitial tissue, and vacuolation of renal tubules were observed in MHV-1 exposed mice kidneys (V) (Figure 9).
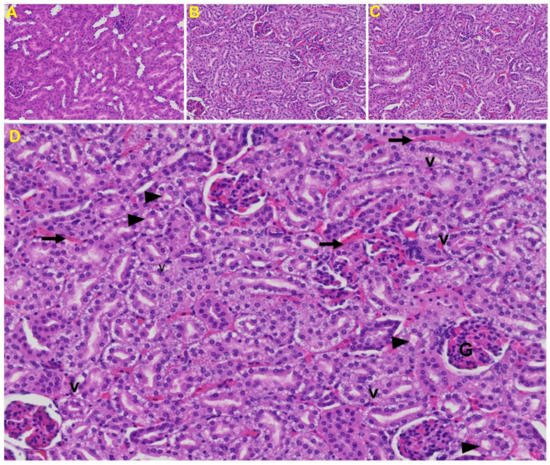
Figure 9. Kidney from an MHV-1 infected mouse. (A) Normal mouse kidney. (B,C) Kidney sections from an MHV-1 infected mouse. Kidney sections from a control mouse showing a normal histological structure with the glomerulus, proximal convoluted tubule, and distal convoluted tubules. (D) Enlarged image of a kidney from an MHV-1 infected mouse showing proximal and distal tubular necrosis (arrowhead), hemorrhage in the interstitial tissue (long arrow), and vacuolation of renal tubules (V) (H&E original magnification of A–C is 400× as noted in Figure 5B).
References
- Kasal, D.A.; De Lorenzo, A.; Tibiriçá, E. COVID-19 and Microvascular Disease: Pathophysiology of SARS-CoV-2 Infection with Focus on the Renin-Angiotensin System. Heart Lung Circ. 2020, 29, 1596–1602.
- Struyf, T.; Deeks, J.J.; Dinnes, J.; Takwoingi, Y.; Davenport, C.; Leeflang, M.M.; Spijker, R.; Hooft, L.; Emperador, D.; Dittrich, S.; et al. Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19 disease. Cochrane Database Syst. Rev. 2020, 7, CD013665, Update in: Cochrane Database Syst. Rev. 2021, 2, CD013665.
- Grant, M.C.; Geoghegan, L.; Arbyn, M.; Mohammed, Z.; McGuinness, L.; Clarke, E.L.; Wade, R.G. The prevalence of symptoms in 24,410 adults infected by the novel coronavirus (SARS-CoV-2; COVID-19): A systematic review and meta-analysis of 148 studies from 9 countries. PLoS ONE 2020, 15, e0234765.
- Damiani, E.; Carsetti, A.; Casarotta, E.; Scorcella, C.; Domizi, R.; Adrario, E.; Donati, A. Microvascular alterations in patients with SARS-COV-2 severe pneumonia. Ann. Intensive Care 2020, 10, 60.
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733.
- Wang, F.; Nie, J.; Wang, H.; Zhao, Q.; Xiong, Y.; Deng, L.; Song, S.; Ma, Z.; Mo, P.; Zhang, Y. Characteristics of Peripheral Lymphocyte Subset Alteration in COVID-19 Pneumonia. J. Infect. Dis. 2020, 221, 1762–1769.
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207.
- Mo, P.; Xing, Y.; Xiao, Y.; Deng, L.; Zhao, Q.; Wang, H.; Xiong, Y.; Cheng, Z.; Gao, S.; Liang, K.; et al. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin. Infect. Dis. 2020, ciaa270.
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513.
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069.
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720.
- Archer, S.L.; Sharp, W.W.; Weir, E.K. Differentiating COVID-19 Pneumonia from Acute Respiratory Distress Syndrome and High Altitude Pulmonary Edema: Therapeutic Implications. Circulation 2020, 142, 101–104.
- Centers for Disease Control and Prevention. Management of Patients with Confirmed 2019-nCoV|CDC. Interim Clinical Guidance for Management of Patients with Confirmed Coronavirus Disease (COVID-19). Available online: http://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-guidance-management-patients.html (accessed on 16 February 2021).
- Johansson, M.A.; Quandelacy, T.M.; Kada, S.; Prasad, P.V.; Steele, M.; Brooks, J.T.; Slayton, R.B.; Biggerstaff, M.; Butler, J.C. SARS-CoV-2 Transmission from People without COVID-19 Symptoms. JAMA Netw. Open. 2021, 4, e2035057.
- Abdool Karim, S.S.; de Oliveira, T. New SARS-CoV-2 Variants—Clinical, Public Health, and Vaccine Implications. N. Engl. J. Med. 2021.
- Forni, D.; Cagliani, R.; Clerici, M.; Sironi, M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017, 25, 35–48.
- Beaudette, E.R.; Hudson, C.B. Cultivation of the virus of infectious bronchitis. J. Am. Vet. Med. Assoc. 1937, 90, 51–58.
- Cheever, F.S.; Daniels, J.B.; Pappenheimer, A.M.; Bailey, O.T. A murine virus (JHM) causing disseminated encephalomyelitis with extensive destruction of myelin: I. Isolation and biological properties of the virus. J. Exp. Med. 1949, 90, 181–194.
- Kahn, J.S.; McIntosh, K. History and recent advances in coronavirus discovery. Pediatr Infect. Dis. J. 2005, 24 (Suppl. S11), S223–S227, discussion S226.
- Tyrrell, D.A.; Bynoe, M.L. Cultivation of viruses from a high proportion of patients with colds. Lancet 1966, 1, 76–77.
- Hamre, D.; Procknow, J.J. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 1966, 121, 190–193.
- Tyrrell, D.A.; Almeida, J.D.; Cunningham, C.H.; Dowdle, W.R.; Hofstad, M.S.; McIntosh, K.; Tajima, M.; Zakstelskaya, L.Y.; Easterday, B.C.; Kapikian, A.; et al. Coronaviridae. Intervirology 1975, 5, 76–82.
- McIntosh, K.; Becker, W.B.; Chanock, R.M. Growth in suckling-mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc. Natl. Acad. Sci. USA 1967, 58, 2268–2273.
- Cheng, V.C.; Lau, S.K.; Woo, P.C.; Yuen, K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007, 20, 660–694.
- Tabibzadeh, A.; Esghaei, M.; Soltani, S.; Yousefi, P.; Taherizadeh, M.; Safarnezhad Tameshkel, F.; Golahdooz, M.; Panahi, M.; Ajdarkosh, H.; Zamani, F.; et al. Evolutionary study of COVID-19, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as an emerging coronavirus: Phylogenetic analysis and literature review. Vet. Med. Sci. 2021, 7, 559–571.
- Forster, P.; Forster, L.; Renfrew, C.; Forster, M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. USA 2020, 117, 9241–9243.
- Li, T.; Liu, D.; Yang, Y.; Guo, J.; Feng, Y.; Zhang, X.; Cheng, S.; Feng, J. Phylogenetic supertree reveals detailed evolution of SARS-CoV-2. Sci. Rep. 2020, 10, 22366.
- Nakagawa, S.; Miyazawa, T. Genome evolution of SARS-CoV-2 and its virological characteristics. Inflamm. Regener. 2020, 40, 17.
- Sahu, A.K.; Sreepadmanabh, M.; Rai, M.; Chande, A. SARS-CoV-2: Phylogenetic origins, pathogenesis, modes of transmission, and the potential role of nanotechnology. Virus Dis. 2021, 32, 1–12.
- Lam, T.T.Y.; Jia, N.; Zhang, Y.W.; Shum, M.H.H.; Jiang, J.F.; Zhu, H.C.; Tong, Y.G.; Shi, Y.X.; Ni, X.B.; Liao, Y.S.; et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 2020, 583, 282–285.
- Worobey, M.; Pekar, J.; Larsen, B.B.; Nelson, M.I.; Hill, V.; Joy, J.B.; Rambaut, A.; Suchard, M.A.; Wertheim, J.O.; Lemey, P. The emergence of SARS-CoV-2 in Europe and North America. Science 2020, 370, 564–570.
- Rehman, S.U.; Shafique, L.; Ihsan, A.; Liu, Q. Evolutionary Trajectory for the Emergence of Novel Coronavirus SARS-CoV-2. Pathogens 2020, 9, 240.
- Wang, J.T.; Lin, Y.Y.; Chang, S.Y.; Yeh, S.H.; Hu, B.H.; Chen, P.J.; Chang, S.C. The role of phylogenetic analysis in clarifying the infection source of a COVID-19 patient. J. Infect. 2020, 81, 147–178.
- Zhang, T.; Wu, Q.; Zhang, Z. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr. Biol. 2020, 30, 1346–1351.e2.
- Hussain, I.; Pervaiz, N.; Khan, A.; Saleem, S.; Shireen, H.; Dong-Qing, W.; Labrie, V.; Bao, Y.; Abbasi, A. Evolutionary and structural analysis of SARS-CoV-2 specific evasion of host immunity. Genes Immun. 2020, 21, 409–419.
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574.
- Dimonaco, N.J.; Salavati, M.; Shih, B.B. Computational Analysis of SARS-CoV-2 and SARS-Like Coronavirus Diversity in Human, Bat and Pangolin Populations. Viruses 2020, 13, 49.
- Junejo, Y.; Ozaslan, M.; Safdar, M.; Khailany, R.A.; Rehman, S.; Yousaf, W.; Khan, M.A. Novel SARS-CoV-2/COVID-19: Origin, pathogenesis, genes and genetic variations, immune responses and phylogenetic analysis. Gene Rep. 2020, 20, 100752.
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123.
- Sagulenko, P.; Puller, V.; Neher, R.A. TreeTime: Maximum-likelihood phylodynamic analysis. Virus Evol. 2018, 4, vex042.
- Jaimes, J.A.; André, N.M.; Chappie, J.S.; Millet, J.K.; Whittaker, G.R. Phylogenetic Analysis and Structural Modeling of SARS-CoV-2 Spike Protein Reveals an Evolutionary Distinct and Proteolytically Sensitive Activation Loop. J. Mol. Biol. 2020, 432, 3309–3325.
- Helmy, Y.A.; Fawzy, M.; Elaswad, A.; Sobieh, A.; Kenney, S.P.; Shehata, A.A. The COVID-19 Pandemic: A Comprehensive Review of Taxonomy, Genetics, Epidemiology, Diagnosis, Treatment, and Control. J. Clin. Med. 2020, 9, 1225.
- Körner, R.W.; Majjouti, M.; Alcazar, M.A.A.; Mahabir, E. Of Mice and Men: The Coronavirus MHV and Mouse Models as a Translational Approach to Understand SARS-CoV-2. Viruses 2020, 12, 880.
- De Albuquerque, N.; Baig, E.; Ma, X.; Zhang, J.; He, W.; Rowe, A.; Habal, M.; Liu, M.; Shalev, I.; Downey, G.P.; et al. Murine hepatitis virus strain 1 produces a clinically relevant model of severe acute respiratory syndrome in A/J mice. J. Virol. 2006, 80, 10382–10394.
- Tian, J.; Milddleton, B.; Kaufman, D.L. GABA administration prevents severe illness and death following coronavirus infection in mice. bioRxiv 2020.
- Marjot, T.; Webb, G.J.; Barritt, A.S., 4th; Moon, A.M.; Stamataki, Z.; Wong, V.W.; Barnes, E. COVID-19 and liver disease: Mechanistic and clinical perspectives. Nat. Rev. Gastroenterol. Hepatol. 2021, 10, 1–17.
- Viner, R.M.; Ward, J.L.; Hudson, L.D.; Ashe, M.; Patel, S.V.; Hargreaves, D.; Whittaker, E. Systematic review of reviews of symptoms and signs of COVID-19 in children and adolescents. Arch. Dis Child. 2020.
- Mohamed, A.A.; Mohamed, N.; Abd-Elsalam, S.; ElSadek, S.M.; Ahmed, H.H.; Taha, H.A.; Mohamed, G.K.; Soliman, D.R. COVID-19 in Pediatrics: A Diagnostic Challenge. Curr. Pediatr. Rev. 2021.
- Loo, J.; Spittle, D.A.; Newnham, M. COVID-19, immunothrombosis and venous thromboembolism: Biological mechanisms. Thorax 2021.
- Price, L.C.; McCabe, C.; Garfield, B.; Wort, S.J. Thrombosis and COVID-19 pneumonia: The clot thickens! Eur. Respir. J. 2020, 56, 2001608.
- Schulman, S.; Hu, Y.; Konstantinides, S. Venous Thromboembolism in COVID-19. Thromb. Haemost. 2020, 120, 1642–1653.
- Bikdeli, B.; Madhavan, M.V.; Jimenez, D.; Chuich, T.; Dreyfus, I.; Driggin, E.; Nigoghossian, C.; Ageno, W.; Madjid, M.; Guo, Y.; et al. COVID-19 and Thrombotic or Thromboembolic Disease: Implications for Prevention, Antithrombotic Therapy, and Follow-Up: JACC State-of-the-Art Review. J. Am. Coll Cardiol. 2020, 75, 2950–2973.
- Suh, Y.J.; Hong, H.; Ohana, M.; Bompard, F.; Revel, M.P.; Valle, C.; Gervaise, A.; Poissy, J.; Susen, S.; Hékimian, G.; et al. Pulmonary Embolism and Deep Vein Thrombosis in COVID-19: A Systematic Review and Meta-Analysis. Radiology 2021, 298, E70–E80.
- Pozzobon, F.M.; Perazzo, H.; Bozza, F.A.; Rodrigues, R.S.; de Mello Perez, R.; Chindamo, M.C. Liver injury predicts overall mortality in severe COVID-19: A prospective multicenter study in Brazil. Hepatol. Int. 2021, 3, 1–9.
- Mukherjee, K.; Banerjee, A.; Bhattacharjee, D.; De, S.; Biswas, A.; Garai, D.; Chakraborty, R.; Manna, A. Liver Function Status in COVID-19: An Indian Perspective. J. Assoc. Physicians India 2021, 69, 19–21.
- Zhao, X.; Lei, Z.; Gao, F.; Xie, Q.; Jang, K.; Gong, J. The impact of coronavirus disease 2019 (COVID-19) on liver injury in China: A systematic review and meta-analysis. Medicine 2021, 100, e24369.
- Wang, Y.; Shi, L.; Wang, Y.; Yang, H. An updated meta-analysis of AST and ALT levels and the mortality of COVID-19 patients. Am. J. Emerg. Med. 2021, 40, 208–209.
- Ramachandran, P.; Perisetti, A.; Gajendran, M.; Chakraborti, A.; Narh, J.T.; Goyal, H. Increased Serum Aminotransferase Activity and Clinical Outcomes in Coronavirus Disease 2019. J. Clin. Exp. Hepatol. 2020, 10, 533–539.
- Deshmukh, V.; Motwani, R.; Kumar, A.; Kumari, C.; Raza, K. Histopathological observations in COVID-19: A systematic review. J. Clin. Pathol. 2021, 74, 76–83.
More
Information
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
953
Entry Collection:
COVID-19
Revisions:
2 times
(View History)
Update Date:
13 Sep 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




