Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Giulia Sancesario | + 4795 word(s) | 4795 | 2021-08-16 05:28:05 | | | |
| 2 | Vivi Li | Meta information modification | 4795 | 2021-09-03 09:51:15 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Sancesario, G. Artificial Intelligence for Alzheimer’s Disease. Encyclopedia. Available online: https://encyclopedia.pub/entry/13857 (accessed on 07 February 2026).
Sancesario G. Artificial Intelligence for Alzheimer’s Disease. Encyclopedia. Available at: https://encyclopedia.pub/entry/13857. Accessed February 07, 2026.
Sancesario, Giulia. "Artificial Intelligence for Alzheimer’s Disease" Encyclopedia, https://encyclopedia.pub/entry/13857 (accessed February 07, 2026).
Sancesario, G. (2021, September 02). Artificial Intelligence for Alzheimer’s Disease. In Encyclopedia. https://encyclopedia.pub/entry/13857
Sancesario, Giulia. "Artificial Intelligence for Alzheimer’s Disease." Encyclopedia. Web. 02 September, 2021.
Copy Citation
The recent growth of open data-sharing initiatives collecting lifestyle, clinical, and biological data from Alzheimer's disease (AD) patients has provided a potentially unlimited amount of information about the disease, far exceeding the human ability to make sense of it. Integrating Big Data from multi-omics studies provides the potential to explore the pathophysiological mechanisms of the entire biological continuum of AD. In this context, Artificial Intelligence (AI) offers a wide variety of methods to analyze large and complex data in order to improve knowledge in the AD field.
Alzheimer’s disease
diagnosis
machine learning
artificial intelligence
1. Introduction
Alzheimer’s disease (AD) is an irreversible neurodegenerative disease that progressively destroys cognitive skills, up to the development of dementia. The clinical diagnosis of AD is based on the presence of objective cognitive deficits (which are, typically, prominent memory impairments). In some cases, AD may show atypical presentations, with impairments in non-amnesic domains (i.e., attention, executive functions, visuo-constructive practice and language) [1]. However, AD shares many common clinical features with other neurodegenerative dementia, including Lewy body dementia [2], frontotemporal disorders [3], and vascular dementia, making early and differential diagnosis difficult, especially in the first stage of the disease [4][5]. Finally, the occurrence of co-existing pathologies is a common feature in those cases of neurodegenerative diseases that share a common pathogenic mechanism, consisting of extracellular and/or intracellular insoluble fibril aggregates of abnormal misfolded proteins (e.g., the formation of amyloid plaques, tau tangles, or α-synuclein inclusions). In this context, the system biology approach, which aims at the integration of clinical and multi-omics data, can help to detect and recognize the pathophysiological and molecular changes characteristic of AD or other pathologies, as well as the associated clinical manifestations occurring, in particular, in the pre-clinical stages [6].
Amyloid plaques and neurofibrillary tangles are the neuropathological hallmarks of the disease [7][8], which can be evaluated in vivo by neuroimaging investigation and cerebrospinal fluid (CSF) biomarker assessment; namely, considering amyloidβ1-42 (Aβ42), its ratio with amyloid β1-40 (Aβ42/Aβ40), total tau protein (t-tau), and hyperphosphorylated tau (p-tau181).
As AD is multi-factorial, many conditions can influence the individual risk and age of onset, particularly metabolic impairments (diabetes mellitus, hypertension, obesity, and low HDL cholesterol), depression, hearing loss, traumatic brain injury, and alcohol abuse [9]. Lifestyle factors such as smoking, low physical activity, and social isolation are potentially modifiable, while several of these may have bi-directional relationships and may be early manifestations, other than risk factors, in the prodromal phase of dementia [10]. All of these clinical, biological, socio-demographic, and lifestyle factors contribute to defining the development of the disease and, therefore, are useful in trying to understand the still-misunderstood etiology of AD.
Decades of experimental and clinical research have contributed to identify many mechanisms in the pathogenesis of the disease, such as the β-amyloid hypothesis. Clinical and biological data from electronic health records and multi-omics sciences represent a potentially unlimited amount of information about biological processes, such as genomes, transcriptomes, and proteomes, which can be explored through Big Data exploitation [6][11]. The rapid collection of data from tens of thousands of AD patients far exceeds the human ability to make sense of the disease.
Complex AI-based models can be successful in extracting meaningful information from Big Data; however, as their complexity increases, it becomes more and more difficult to interpret how they produce their output. Thus, they have been called black-box models. Making AI explainable is a key problem of AI technological development in recent years, and is of pivotal importance in healthcare applications, where both patients and clinicians need to trust research methods to make decisions about people’s health [12]. The AD pathology is characterized by high complexity and heterogeneity, and many authors have demonstrated the absence of etiological uniformity and diverse treatment suitability for each patient, supporting the need for an accurate individual diagnosis [13][14]. In order to make the most of biological experiments and refine their findings, they have to be supported and followed by complex biological modeling, based on mathematical and statistical tools such as Artificial Neural Networks [15]. Formalized domain expertise from psychology, neuroscience, neurology, psychiatry, geriatric medicine, biology, and genetics can be integrated with novel analytic approaches from bioinformatics and statistics to be applied on Big Data in AD research projects, with the aim of answering detailed questions through the use of predictive models. These can succeed in answering key questions about promising biomarker combinations, patient sub-groups, and disease progression, finally leading to the development of effective treatment strategies, helping patients with tailored medical approaches [16].
In this context, AI technology represents a promising approach to investigate the pathological mechanisms of AD by analyzing such complex data. In this review, we focus on recent findings using AI for AD research and future challenges awaiting its application: Will it be possible to make an early diagnosis of AD with AI? Will AI be able to predict conversion from Mild Cognitive Impairment (MCI) to AD dementia, stratify patients, and identify “malignant” forms with worse disease progression? Finally, will AI be able to predict the course and progression of the disease and help in finding a cure for AD?
1.1. AI and the Biomedical Research
AI has recently significantly revolutionized the way that digital data is analyzed and used. Currently, in some applications, AI is used to perform simple tasks, such as face or speech recognition, and often outperforms human abilities in those tasks [17]. This is definitely a great opportunity to be transferred to medical care, due to the potential for fast, low-cost, and accurate automation; for example, in the processing of digital images by AI algorithms [18]. Several studies have been performed with the attempt to ameliorate the knowledge of complex multifactorial diseases such as AD: AI exploits the features of Machine Learning (ML) and Deep Learning (DL) to develop algorithms that can be used in the clinical and biomedical fields for the classification and stratification of patients, based on the integration and processing of a large variety of available data sources, including neuroimaging, biochemical markers, clinical, and neuropsychological (NPS) data from patients and controls. An extensive application of AI in the biomedical field is for Computer-Aided Diagnosis (CAD). This kind of application aims to automate the diagnostic process upon data analysis, potentially contributing to reaching an early and differential diagnosis of AD or dementia of different etiology (Figure 1).
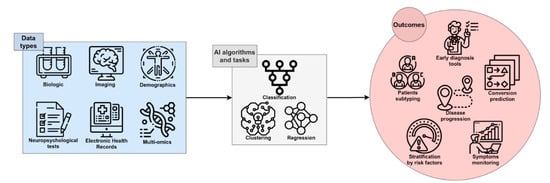
Figure 1. The framework of AI in AD research. Single-modality or integrated data can be processed by several algorithms and tasks, leading to useful outcomes for early and accurate diagnosis, prediction of the course and progression of disease, patient stratification, and discovery of novel therapeutic targets and disease-modifying therapies.
ML is a fundamental branch of AI, consisting of a collection of data analysis techniques that aim to generate predictive models by learning from data, progressively improving the ability to make predictions through experience (see glossary; Table 1) [19]. DL is a sub-field of ML and uses methods that are able to learn relationships between inputs and outputs by modeling highly non-linear interactions into higher representations, at a more abstract level (see glossary) [20]. Moreover, there are two main categories of predictive models based on ML or DL techniques: Supervised and unsupervised. In supervised learning, the algorithms learn from labeled data to associate an input (e.g., cortical thicknesses data) with a specific output (e.g., presence/absence of a disease or neuropsychological test performance), leading to models that are able to predict the output variable. In unsupervised learning, the algorithms learn from unlabeled data with the purpose of identifying clusters among the observations, based on similar features (see glossary); unlike in supervised learning, there are no correct answers and the aim of the algorithm is to discover structures within variables.
Table 1. Glossary.
| Method | Definition | Details |
|---|---|---|
| Machine Learning | A collection of data analysis techniques that aim to generate predictive models by learning from data and improving their ability to make predictions through experience. | ML models are considered shallow learners, working on data with hand-crafted features defined through expert-based knowledge. Raw data must be pre-processed before constructing a ML system, requiring domain expertise to proceed with feature extraction and engineering, in order to train the algorithm appropriately. As an example of a ML algorithm, a Support Vector Machine (SVM) accomplishes the classification task by finding the hyperplane that, in the multi-dimensional feature space, optimally separates the data into two (or more) classes. |
| Deep Learning | A sub-field of ML that uses methods that are able to learn relationships between inputs and outputs by modeling highly non-linear interactions. | DL models are different from shallow learners and can elaborate raw data, thus requiring little or no feature engineering, thanks to their ability to model complex functions and identify relevant aspects in the data distribution. DL algorithms are based on Artificial Neural Networks (ANNs), which are inspired by the human brain and can model very complex functions, identifying important aspects in the features and suppressing irrelevant ones. As an example of a DL algorithm, a Convolutional Neural Network (CNN) is composed of nodes organized into layers. It can take an image as input, elaborate the features of the image through its layers, and assign a class attribution as output, thus differentiating between two or more groups. |
| Supervised learning | A ML task defined through the use of labeled data sets for algorithm training. | The algorithms learn to give the right answer, as defined by the ground truth set, which has labels assigned to the data.An SVM performing the classification task is an example of an algorithm trained by supervised learning. |
| Unsupervised learning | A set of algorithms aimed to discover hidden patterns or data groupings without the need for human intervention. | In unsupervised learning, unlike supervised learning, there are no correct answers and the algorithm’s aim is to discover structures within variables. The algorithms work with unlabeled data.Common unsupervised methods include clustering algorithms and Principal Component Analysis (PCA). |
| Classification task | The algorithm is trained to predict a class label. | A classical example is the classification of patients affected by a disease vs. normal controls. The algorithm learns to associate input data with an output label in a supervised manner, and its results can be evaluated by metrics such as accuracy score. |
| Regression task | The algorithm is trained to predict the value of a continuous variable. | An example is the prediction of hippocampus volume as a numerical quantity. The algorithm learns to associate input data with an output value in a supervised manner, and its results can be evaluated by metrics such as the Root Mean Squared Error (RMSE). |
| Clustering | Clustering consists of partitioning a data set, in order to find a grouping of the data points. | Clustering is one of the most important unsupervised learning techniques. Its main goal is to reveal sub-groups within heterogeneous data, in such a way that greater homogeneity is shown within clusters (rather than between clusters). Clustering algorithms can lead to the identification of patterns across subjects or patients that are difficult to find even for an expert clinician. |
| Overfitting | Overfitting occurs when the model is too dependent on training data to make accurate predictions on test data. | When a model is overfitted, its learned ability to separate between two classes does not generalize well to data it has never seen before, therefore limiting its usability for real-world applications. |
| Ensemble learning | Model ensembling consists of combining multiple ML models, in order to obtain better predictive performance than any of the constituents alone. | A single model alone can be weak in generating predictions. Combining multiple models can compensate for their individual weaknesses. |
| Transfer learning | A supervised learning technique in which the knowledge previously acquired from the model in one task is used to solve related ones. | In a transfer learning approach, the model is first pre-trained on a source task, then re-trained and tested on a target task. The source task should be related to the target task, with similar relations between the input and output data. In fact, in the pre-training phase, the model gains helpful knowledge for the target task. |
| Cox regression | The Cox proportional hazards model is a regression technique for investigating the association between the time of an event occurring and one or more predictor variables. | Cox regression gives hazard rates as measures of how factors influence the risk for an event occurrence (outcome), be it death or infection. |
AI techniques have found a wide range of applications in the clinical and biomedical fields, in an effort to automate, standardize, and improve the accuracy of early prediction (regression task), classification of patients (classification task), or the stratification of subjects, based on the processing of specific data (see glossary). In a classification task, the algorithm is trained to associate a label (e.g., “AD diagnosis”) to a given set of features (e.g., clinical parameters, cognitive status, genotypes, biochemical markers, imaging, and so on), in order to be able to generate predictions. Once the model is ready, it is able to predict a class, defined by a label (e.g., “AD” or “control”), by analyzing the set of features of a new given example. The regression task instead involves predicting the value of a variable (e.g., “hippocampus volume” or “biomarker level”) measured on a continuous scale.
Despite ample research effort, we still do not have a cure capable of modifying and/or halting the course of the disease. Some clinical trials are ongoing, especially with the use of monoclonal antibodies targeting Aβ peptides, modified Aβ species, and monomeric as well as aggregated oligomers, which have shown to be safe and have clinical efficacy in AD patients [21]. However, AI pipelines can be applied in automatic compound synthesis in order to analyze the literature and high-throughput compound screening data, to perform an initial molecular screening and automated chemical synthesis [22]. By updating the AI model after cell- or organoid-based experiments, AI can be used to propose a new molecular optimization plan and new bioassays can be conducted to evaluate the biological effects of the compound, thus enabling an automated drug development cycle based on AI design and high-throughput bioassay, greatly accelerating the development of new drugs [23]. AI technology can be used to repurpose known drugs for treatment of Alzheimer’s disease [22][24][25][26]. This is a fast, low-cost drug development pathway, in which AI is used to predict drug repurposing by analyzing large-scale transcriptomics, molecular structure data, and clinical databases. Finally, AI can be exploited to simplify clinical trials too, both in the design and implementation phase [22]. Participant selection can be optimized by using AI algorithms on genetic and clinical data, thus predicting which subset of the population may be sensitive to new drugs [27]. Notably, coupling AI with data from wearables enables almost real-time non-invasive diagnostics, potentially preventing drop-out at subject level [28]. Although promising and rapidly growing, only few of these AI applications have made it to the clinical application stage; nonetheless, AI represents a promising technology to support research and, finally, to develop novel effective therapies [22][29].
1.2. Public Databases and Biobanks
The application of AI-based techniques for AD and other diseases research requires extensive data sets, composed of hundreds to thousands of entries describing subjects over many clinical and biological variables, which can be employed to develop novel algorithms by analyzing the features of the disease. In the last 20 years, many open data-sharing initiatives have grown in the field of neurodegenerative disease research [30]; and, in particular, in AD research. Some important data-sharing resources are the Alzheimer’s Disease Genetics Consortium (ADGC, www.adgenetics.org, accessed on 30 May 2021 Date Month Year), Alzheimer’s Disease Sequencing Project (ADSP, www.niagads.org/adsp/content/home accessed on 30 May 2021), Alzheimer’s Disease Neuroimaging Initiative (ADNI, http://adni.loni.usc.edu/ accessed on 30 May 2021), AlzGene (www.alzgene.org accessed on 30 May 2021), Dementias Platform UK (DPUK, https://portal.dementiasplatform.uk/ accessed on 30 May 2021), Genetics of Alzheimer’s Disease Data Storage Site (NIAGADS, www.niagads.org/ accessed on 30 May 2021), Global Alzheimer’s Association Interactive Network (GAAIN, www.gaain.org/ accessed on 30 May 2021), and National Centralized Repository for Alzheimer’s Disease and Related Dementias (NCRAD, https://ncrad.iu.edu accessed on 30 May 2021) [31][32]. Such public databases and repositories collect biological specimens and data from clinical and cognitive tests; lifestyle, neuroimages; genetics; and CSF and blood biomarkers from normal, cognitively impaired, or demented individuals, which can be combined to apply cutting-edge ML algorithms. Moreover, the National Alzheimer’s Coordinating Center (NACC) has constructed a large relational database for both exploratory and explanatory AD research, by use of standardized clinical and neuropathological research data [33]; DementiaBank, the component of TalkBank dedicated to data on language in dementia, provides data sets from verbal tasks such as the Pitt corpus, which contain audio files and text transcriptions from AD subjects and controls [34][35].
In the so-called “Omics era”, several databases of omics data—not limited to or specific for neurodegenerative diseases—have been established, such as the Gene Expression Omnibus (GEO), which collects functional genomics data of array- and sequence-based data regarding many physiological and pathological conditions, including AD, among others [36]. Finally, UK Biobank collects and stores healthcare databases and associated biological specimens for a wide range of health-related outcomes from a large prospective study including over 500,000 participants [37].
In the AD field, public and private databases represent the substrate and the source for AI to facilitate a more comprehensive understanding of disease heterogeneity, as well as personalized medicine and drug development.
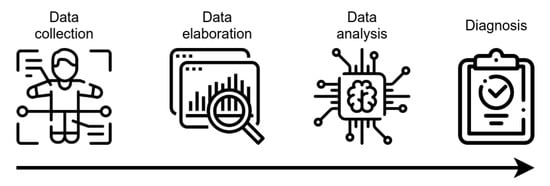
2. AI for AD Diagnosis
AI technology, mainly ML algorithms, can handle high-dimensional complex systems that exceed the human capacity of data analysis. ML has been used in the CAD of many pathologies, including AD, by combining electronic medical records, NPS tests, brain imaging, and biological markers, together with data obtained by novel developed tools (e.g., wearable sensors) for the assessment of executive functions (Figure 2). Magnetic Resonance Imaging (MRI), Positron Emission Tomography (PET), 18F-fluorodeoxyglucose-Positron Emission Tomography (FDG-PET), and Diffusion Tensor Imaging (DTI) provide detailed information about the brain structure and functionality, allowing for the identification of features supporting the diagnosis, such as atrophy, amyloid deposition, or microstructural damages [38][39]. Moreover, neuroimages can discriminate pathological processes not due to AD that can lead to cognitive decline (e.g., brain tumors or cerebrovascular disease). Several studies have demonstrated that markers of primary AD pathology (CSF Aβ1-42, total tau and p-tau181, amyloid-PET), neurodegeneration (structural MRI, FDG-PET), or biomarker combinations can be integrated into complex tools for diagnostic or predictive purposes [40][41][42][43]. Of interest, polymorphism in the apolipoprotein E (APOE) gene is the strongest genetic risk factor in the sporadic form of AD, which has an added predictive value, with the APOEε4 allele conferring an increased risk of early age of onset, while the APOEε2 allele confers a decreased risk, relative to the common APOEε3 allele [44].

Figure 2. Schematic representation of CAD tools functioning. After collection, data are elaborated, in order to be made ready for the analysis using AI-based techniques. The outcoming result is the assignment of a class, with potential value for diagnostic evaluation.
So far, the first CAD tools for AD were constructed through the use of AI methods for the analysis of brain imaging [45][46]. Analyzing MRI data from the OASIS database [47], a feature extraction and selection method called “eigenbrain”, which is carried out using PCA (see glossary), was used to capture the characteristic changes of anatomical structures between AD and NC; namely, severe atrophy of the cerebral cortex, enlargement of the ventricles, and shrinkage of the hippocampus. The applied SVM algorithm achieved a mean accuracy of 92.36% for an automated classification system of AD diagnosis, based on MRI data [48]. For instance, by using FDG-PET of the brain, a DL algorithm for the early prediction of AD was developed, achieving 82% specificity and 100% sensitivity at an average of 75.8 months prior to the final diagnosis [49].
The majority of ML models for classifying AD from NC are trained with neuroimaging data, which have the advantage of high accuracy [50], but limitations associated to their high cost and lack of diffusion in non-specialized centers. A large group of studies have focused on the identification of fluid marker panels as potential screening tests. In addition to Aβ- and tau-related biomarkers, novel candidate markers according to other mechanisms of AD pathology have been investigated in experimental and meta-analysis studies, in order to optimize the predictive modeling.
A recent study has applied ML algorithms to evaluate data on novel biomarkers that were available on PubMed, Cochrane Systematic Reviews, and Cochrane Collaboration Central Register of Controlled Clinical Trials databases. Experimental or review studies have investigated biomarkers for dementia or AD using ML algorithms, including SVM, logistic regression, random forest, and naïve Bayes. The panel included indices of synaptic dysfunction and loss, neuroinflammation, and neuronal injury (e.g., neurofilament light; NFL). An algorithm, developed by integrating all the data from such fluid biomarkers, has been shown to be capable of accurately predicting AD, thus achieving state-of-the-art results [51].
To the end of designing a blood-based test for identifying AD, the European Medical Information Framework for Alzheimer’s disease biomarker discovery cohort conducted a study using both ML and DL models. Data used for modeling included 883 plasma metabolites assessed in 242 cognitively normal individuals and 115 patients with AD-type dementia, and demonstrated that the panel of plasma markers had good discriminatory power and have the potential to match the AUC of well-established AD CSF biomarkers. Finally, the authors concluded that it can be commonly included in clinical research as part of the diagnostic work-up, with an AUC in the range of 0.85–0.88 [52].
Moreover, AI has the potential to integrate data obtained through the use of new technologies, such as devices designed for the evaluation of language and verbal fluency or executive functions in healthy or mildly impaired individuals. A system for acoustic feature extraction over speech segments in AD patients was developed, by analyzing data from DementiaBank’s Pitt corpus data set [34][35]. The acoustic features of patient speeches have the advantage of being cost-effective and non-invasive, compared to imaging or blood biomarkers, and can be integrated to develop screening tools for MCI and AD. Moreover, another system has exploited a digital pen to record drawing dynamics, which can detect slight signs of mild impairment in asymptomatic individuals [53]. The ability of this pen was evaluated in patients performing the Clock Drawing Test (CDT), which allowed for the identification of subtle to mild cognitive impairment, with an inexpensive and efficient tool having promising clinical and pre-clinical applications.
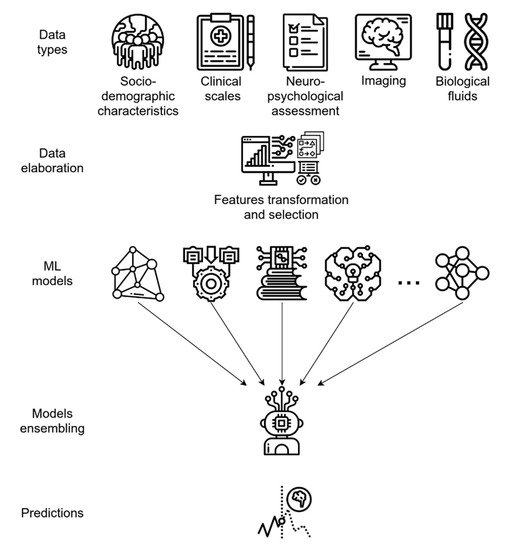
3. Prediction of MCI-to-AD Conversion
Diagnosing probable AD in a subject with moderate–severe cognitive decline or evidence of cortex atrophy is usually not difficult for a skilled neurologist when appropriate data are available. Therefore, it is not surprising for an AI model to solve the task of AD vs. NC subject classification with high accuracy, when taking into account NPS test results or neuroimaging data [54]. To date, several predictive models have been developed [54][55][56][57][58][59][60][61][62][63][64][65][66][67][68][69][70][71], yielding peak accuracy values of 100% in AD vs. NC classification [54]. In contrast, a much more challenging task for AI is to identify individuals with subjective or mild impairment who will develop AD dementia, with respect to stable MCI or MCI not due to AD, given the shaded differences and the overlapping symptoms in the clinical or biological variables defining these groups in the early phases [72].
Algorithms designed to predict MCI-to-AD conversion aim to classify MCI patients into two groups: those who will convert to AD (MCI-c) within a certain time frame (usually 3 years) and those who will not convert (MCI-nc). Yearly, about 15% of MCI patients convert to AD [59][73] and, thus, early and timely identification is crucial, in order to ameliorate the outcome and slow the progression of the disease.
Several AI-based models test the accuracy of combinations of non-invasive predictors, as well as socio-demographic and clinical data, in order to develop effective screening or predictive tools.
By using the ADNI data set, socio-demographic characteristics, clinical scale ratings, and NPS test scores have been used to train different supervised ML algorithms and, finally, develop an ensemble model utilizing them (see glossary). This ensemble learning application demonstrated a high predictive performance, with an AUC of 0.88 in predicting MCI-to-AD conversion [61], and has the advantage of using only non-invasive and easily collectable predictors, rather than neuroimaging or CSF biomarkers, thus enhancing its potential use and diffusion in clinical practice.
As for CAD systems, both MRI and PET data can be independently modeled by ML algorithms, yielding good predictive accuracy; however, integrating neuroimaging data with other variables, such as cognitive measures, genetic factors, or biochemical changes, can significatively enhance the model performance, as is generally expected when integrating multi-modal data (Figure 3) [30][63][65][70][71]. For example, the integration of MRI with multiple modality data, such as PET, CSF biomarkers, and genomic data, reached 84.7% accuracy in an MCI-c vs. MCI-nc classification task. When only single-modality data was used, the accuracy of the model was lower than the all-modalities implementation [63].

Figure 3. Predictive ML ensemble method for the conversion of MCI to AD based on multi-modal data (i.e., socio-demographics, clinical, NPS, biological fluids, and imaging data). The system uses a feature transformation and selection phase followed by data integration, allowing for more efficient use of variables. The final ensemble of several different ML models provides accurate final predictions of AD or AD conversion.
Different data modalities reflect the AD-related pathological markers that are complementary to each other, which can be concatenated as multi-modal features as input to an ML model for classification [74][75][76]. Notwithstanding, the modality with the larger number of features may weigh more than the others when training the algorithm, inducing bias in the interpretation. In order to overcome this limitation and extract multi-modal feature representations, DL architectures can be used, which do not need feature engineering, due to their ability to non-linearly transform input variables [77].
A DL model for both MCI-to-AD prediction and AD vs. NC classification was trained on data from the ADNI database, including demographic, NPS and genetic data, APOE polymorphism, and MRI. The model processed all the data in a multi-modal feature extraction phase, aiming to combine all data together and obtain a classification output. The AD vs. NC classification task achieved by this model reached performances close to 100%, as expected; whereas, for the MCI-to-AD prediction task, the AUC and accuracy were 0.925 and 86%, respectively [54].
Some models transferred the knowledge in performing AD vs. NC classification to a prediction task—that is, MCI-to-AD conversion—using transfer learning methodology (see glossary). A system with the highest capacity of discriminating AD from NC by analyzing three-dimensional MRI data was recently tested for MCI-to-AD prediction, reaching a high accuracy (82.4%) and AUC (0.83). This finding demonstrated that information from related domains can help AI to solve tasks targeted at the identification of patients at risk of developing AD-related dementia [55].
An interpretation system was embedded with a classification model for both early diagnosis of AD and MCI-to-AD prediction, in order to increase the impact in clinical practice. This model integrates 11 data modalities from ADNI, including NPS tests, neuroimaging, demographics, and electronic health records data (e.g., laboratory blood test, neurological exam, and clinical symptom data). The model outputs a sentence in natural language, explaining the involvement of attributes in the model’s classification output. The model achieved a good performance while balancing the accuracy–interpretability trade-off in both AD classification and MCI-to-AD prediction tasks, allowing for actionable decisions that can enhance physician confidence, contributing to the realization of explainable AI (XAI) in healthcare [78].
Most of the AI models for AD are mainly built on biomarkers such as brain imaging, often with the use of Aβ and tau ligands, Aβ- or tau-PET, as well as biomarkers in CSF, which have high accuracy and predictive value; however, their invasive nature, high cost, and limited availability restrict their use to highly specialized centers [79][80][81][82]. A possible turning point has emerged with the recent development of ultra-sensitive methods for the detection of brain-derived proteins in blood, making it possible to measure NFL [83], Aβ42, and Aβ40 [84][85], and tau and P-tau in plasma [86][87]. The accuracy of plasma P-tau combined with other non-invasive biomarkers for predicting future AD dementia was recently evaluated in patients with mild cognitive symptoms from ADNI and the Swedish BioFINDER cohort, including patients with repeated examinations and clinical assessments over a period of 4 years to ensure a clinical diagnosis (https://biofinder.se/ accessed on 30 May 2021). The prediction included not only the discrimination between progression to AD dementia and stable cognitive symptoms, but also versus progression to other forms of dementia. The accuracy of plasma biomarkers was compared with corresponding markers in CSF, and with the diagnostic prediction of expert physicians in memory clinics, based on the assessment at baseline of extensive clinical assessments, cognitive testing, and structural brain imaging [87]. Plasma P-tau in combination with the other non-invasive markers showed a higher value in predicting AD dementia within 4 years, with respect to clinical-based prediction (AUC of 0.89–0.92 and 0.72, respectively). In addition, the biomarker combination showed similarly high predictive accuracy in both plasma and CSF, making plasma an effective alternative to CSF, thus providing a tool to improve the diagnostic potential in clinical practice [87].
References
- Knopman, D.S.; Amieva, H.; Petersen, R.C.; Chételat, G.; Holtzman, D.M.; Hyman, B.T.; Nixon, R.A.; Jones, D.T. Alzheimer Disease. Nat. Rev. Dis. Primers 2021, 7, 33.
- McKeith, I.G.; Boeve, B.F.; Dickson, D.W.; Halliday, G.; Taylor, J.-P.; Weintraub, D.; Aarsland, D.; Galvin, J.; Attems, J.; Ballard, C.G.; et al. Diagnosis and Management of Dementia with Lewy Bodies: Fourth Consensus Report of the DLB Consortium. Neurology 2017, 89, 88–100.
- Rascovsky, K.; Hodges, J.R.; Knopman, D.; Mendez, M.F.; Kramer, J.H.; Neuhaus, J.; van Swieten, J.C.; Seelaar, H.; Dopper, E.G.P.; Onyike, C.U.; et al. Sensitivity of Revised Diagnostic Criteria for the Behavioural Variant of Frontotemporal Dementia. Brain 2011, 134, 2456–2477.
- Beach, T.G.; Monsell, S.E.; Phillips, L.E.; Kukull, W. Accuracy of the Clinical Diagnosis of Alzheimer Disease at National Institute on Aging Alzheimer’s Disease Centers, 2005–2010. J. Neuropathol. Exp. Neurol. 2012, 71, 266–273.
- Sancesario, G.M.; Bernardini, S. Diagnosis of Neurodegenerative Dementia: Where Do We Stand, Now? Ann. Transl. Med. 2018, 6, 340.
- Hampel, H.; Nisticò, R.; Seyfried, N.T.; Levey, A.I.; Modeste, E.; Lemercier, P.; Baldacci, F.; Toschi, N.; Garaci, F.; Perry, G.; et al. Omics Sciences for Systems Biology in Alzheimer’s Disease: State-of-the-Art of the Evidence. Ageing Res. Rev. 2021, 69, 101346.
- Braak, H.; Braak, E. Neuropathological Stageing of Alzheimer-Related Changes. Acta Neuropathol. 1991, 82, 239–259.
- Braak, H.; de Vos, R.A.; Jansen, E.N.; Bratzke, H.; Braak, E. Neuropathological Hallmarks of Alzheimer’s and Parkinson’s Diseases. Prog. Brain Res. 1998, 117, 267–285.
- Livingston, G.; Huntley, J.; Sommerlad, A.; Ames, D.; Ballard, C.; Banerjee, S.; Brayne, C.; Burns, A.; Cohen-Mansfield, J.; Cooper, C.; et al. Dementia Prevention, Intervention, and Care: 2020 Report of the Lancet Commission. Lancet 2020, 396, 413–446.
- Singh-Manoux, A.; Dugravot, A.; Fournier, A.; Abell, J.; Ebmeier, K.; Kivimäki, M.; Sabia, S. Trajectories of Depressive Symptoms Before Diagnosis of Dementia. JAMA Psychiatry 2017, 74, 712–718.
- Sancesario, G.M.; Bernardini, S. Alzheimer’s Disease in the Omics Era. Clin. Biochem. 2018, 59, 9–16.
- Gunning, D.; Stefik, M.; Choi, J.; Miller, T.; Stumpf, S.; Yang, G.-Z. XAI—Explainable Artificial Intelligence. Sci. Robot. 2019, 4.
- Quinn, J.F.; Raman, R.; Thomas, R.G.; Yurko-Mauro, K.; Nelson, E.B.; Van Dyck, C.; Galvin, J.E.; Emond, J.; Jack, C.R.; Weiner, M. Docosahexaenoic Acid Supplementation and Cognitive Decline in Alzheimer Disease: A Randomized Trial. JAMA 2010, 304, 1903–1911.
- Sepulveda-Falla, D.; Chavez-Gutierrez, L.; Portelius, E.; Vélez, J.I.; Dujardin, S.; Barrera-Ocampo, A.; Dinkel, F.; Hagel, C.; Puig, B.; Mastronardi, C. A Multifactorial Model of Pathology for Age of Onset Heterogeneity in Familial Alzheimer’s Disease. Acta Neuropathol. 2021, 141, 217–233.
- Richards, B.A.; Lillicrap, T.P.; Beaudoin, P.; Bengio, Y.; Bogacz, R.; Christensen, A.; Clopath, C.; Costa, R.P.; de Berker, A.; Ganguli, S. A Deep Learning Framework for Neuroscience. Nat. Neurosci. 2019, 22, 1761–1770.
- Geerts, H.; Dacks, P.A.; Devanarayan, V.; Haas, M.; Khachaturian, Z.S.; Gordon, M.F.; Maudsley, S.; Romero, K.; Stephenson, D.; Initiative, B.H.M. Big Data to Smart Data in Alzheimer’s Disease: The Brain Health Modeling Initiative to Foster Actionable Knowledge. Alzheimer’s Dement. 2016, 12, 1014–1021.
- Vinyals, O.; Babuschkin, I.; Czarnecki, W.M.; Mathieu, M.; Dudzik, A.; Chung, J.; Choi, D.H.; Powell, R.; Ewalds, T.; Georgiev, P.; et al. Grandmaster Level in StarCraft II Using Multi-Agent Reinforcement Learning. Nature 2019, 575, 350–354.
- American Medical Association Augmented Intelligence in Health Care. 2019. Available online: https://www.ama-assn.org/system/files/2019-01/augmented-intelligence-policy-report.pdf (accessed on 30 April 2021).
- Mitchell, T.M. Machine Learning; McGraw-Hill series in computer science; McGraw-Hill: New York, NY, USA, 1997; ISBN 978-0-07-042807-2.
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444.
- Tolar, M.; Hey, J.; Power, A.; Abushakra, S. Neurotoxic Soluble Amyloid Oligomers Drive Alzheimer’s Pathogenesis and Represent a Clinically Validated Target for Slowing Disease Progression. Int. J. Mol. Sci. 2021, 22, 6355.
- Mishra, R.; Li, B. The Application of Artificial Intelligence in the Genetic Study of Alzheimer’s Disease. Aging Dis. 2020, 11, 1567–1584.
- Schneider, G. Automating Drug Discovery. Nat. Rev. Drug Discov. 2018, 17, 97–113.
- Zhang, M.; Schmitt-Ulms, G.; Sato, C.; Xi, Z.; Zhang, Y.; Zhou, Y.; St George-Hyslop, P.; Rogaeva, E. Drug Repositioning for Alzheimer’s Disease Based on Systematic ‘Omics’ Data Mining. PLoS ONE 2016, 11, e0168812.
- Kumar, S.; Chowdhury, S.; Kumar, S. In Silico Repurposing of Antipsychotic Drugs for Alzheimer’s Disease. BMC Neurosci. 2017, 18, 76.
- Xu, Y.; Kong, J.; Hu, P. Computational Drug Repurposing for Alzheimer’s Disease Using Risk Genes from GWAS and Single-Cell RNA Sequencing Studies. Front. Pharmacol. 2021, 12, 617537.
- Harrer, S.; Shah, P.; Antony, B.; Hu, J. Artificial Intelligence for Clinical Trial Design. Trends Pharmacol. Sci. 2019, 40, 577–591.
- He, J.; Baxter, S.L.; Xu, J.; Xu, J.; Zhou, X.; Zhang, K. The Practical Implementation of Artificial Intelligence Technologies in Medicine. Nat. Med. 2019, 25, 30–36.
- Tran, B.X.; Vu, G.T.; Ha, G.H.; Vuong, Q.-H.; Ho, M.-T.; Vuong, T.-T.; La, V.-P.; Ho, M.-T.; Nghiem, K.-C.P.; Nguyen, H.L.T. Global Evolution of Research in Artificial Intelligence in Health and Medicine: A Bibliometric Study. J. Clin. Med. 2019, 8, 360.
- Termine, A.; Fabrizio, C.; Strafella, C.; Caputo, V.; Petrosini, L.; Caltagirone, C.; Giardina, E.; Cascella, R. Multi-Layer Picture of Neurodegenerative Diseases: Lessons from the Use of Big Data through Artificial Intelligence. J. Pers. Med. 2021, 11, 280.
- Ashish, N.; Bhatt, P.; Toga, A.W. Global Data Sharing in Alzheimer Disease Research. Alzheimer Dis. Assoc. Disord. 2016, 30, 160–168.
- Bertram, L.; McQueen, M.B.; Mullin, K.; Blacker, D.; Tanzi, R.E. Systematic meta-analyses of Alzheimer disease genetic association studies: The AlzGene database. Nat. Genet. 2007, 39, 17–23.
- Beekly, D.L.; Ramos, E.M.; Lee, W.W.; Deitrich, W.D.; Jacka, M.E.; Wu, J.; Hubbard, J.L.; Koepsell, T.D.; Morris, J.C.; Kukull, W.A.; et al. The National Alzheimer’s Coordinating Center (NACC) Database: The Uniform Data Set. Alzheimer Dis. Assoc. Disord. 2007, 21, 249–258.
- Haider, F.; de la Fuente, S.; Luz, S. An Assessment of Paralinguistic Acoustic Features for Detection of Alzheimer’s Dementia in Spontaneous Speech. IEEE J. Sel. Top. Signal Process. 2020, 14, 272–281.
- Becker, J.T.; Boiler, F.; Lopez, O.L.; Saxton, J.; McGonigle, K.L. The Natural History of Alzheimer’s Disease: Description of Study Cohort and Accuracy of Diagnosis. Arch. Neurol. 1994, 51, 585–594.
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucleic Acids Res. 2002, 30, 207–210.
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med. 2015, 12, e1001779.
- Spalletta, G.; Piras, F.; Piras, F.; Sancesario, G.; Iorio, M.; Fratangeli, C.; Cacciari, C.; Caltagirone, C.; Orfei, M.D. Neuroanatomical Correlates of Awareness of Illness in Patients with Amnestic Mild Cognitive Impairment Who Will or Will Not Convert to Alzheimer’s Disease. Cortex 2014, 61, 183–195.
- Giulietti, G.; Torso, M.; Serra, L.; Spanò, B.; Marra, C.; Caltagirone, C.; Cercignani, M.; Bozzali, M. Alzheimer’s Disease Neuroimaging Initiative (ADNI) Whole Brain White Matter Histogram Analysis of Diffusion Tensor Imaging Data Detects Microstructural Damage in Mild Cognitive Impairment and Alzheimer’s Disease Patients. J. Magn. Reson. Imaging 2018.
- Albert, M.S.; DeKosky, S.T.; Dickson, D.; Dubois, B.; Feldman, H.H.; Fox, N.C.; Gamst, A.; Holtzman, D.M.; Jagust, W.J.; Petersen, R.C.; et al. The Diagnosis of Mild Cognitive Impairment due to Alzheimer’s Disease: Recommendations from the National Institute on Aging-Alzheimer’s Association Workgroups on Diagnostic Guidelines for Alzheimer’s Disease. Alzheimer’s Dement. 2011, 7, 270–279.
- McKhann, G.M.; Knopman, D.S.; Chertkow, H.; Hyman, B.T.; Jack, C.R.; Kawas, C.H.; Klunk, W.E.; Koroshetz, W.J.; Manly, J.J.; Mayeux, R.; et al. The Diagnosis of Dementia due to Alzheimer’s Disease: Recommendations from the National Institute on Aging-Alzheimer’s Association Workgroups on Diagnostic Guidelines for Alzheimer’s Disease. Alzheimer’s Dement. 2011, 7, 263–269.
- Sperling, R.A.; Aisen, P.S.; Beckett, L.A.; Bennett, D.A.; Craft, S.; Fagan, A.M.; Iwatsubo, T.; Jack, C.R.; Kaye, J.; Montine, T.J.; et al. Toward Defining the Preclinical Stages of Alzheimer’s Disease: Recommendations from the National Institute on Aging-Alzheimer’s Association Workgroups on Diagnostic Guidelines for Alzheimer’s Disease. Alzheimer’s Dement. 2011, 7, 280–292.
- Jack, C.R.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a Biological Definition of Alzheimer’s Disease. Alzheimer’s Dement. 2018, 14, 535–562.
- Williamson, J.; Goldman, J.; Marder, K.S. Genetic Aspects of Alzheimer Disease. Neurologist 2009, 15, 80–86.
- Salas-Gonzalez, D.; Górriz, J.M.; Ramírez, J.; López, M.; Álvarez, I.; Segovia, F.; Puntonet, C.G. Computer Aided Diagnosis of Alzheimer Disease Using Support Vector Machines and Classification Trees. In Advances in Neuro-Information Processing; Köppen, M., Kasabov, N., Coghill, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 418–425.
- Gorriz, J.M.; Ramirez, J.; Lassl, A.; Salas-Gonzalez, D.; Lang, E.W.; Puntonet, C.G.; Alvarez, I.; Lopez, M.; Gomez-Rio, M. Automatic Computer Aided Diagnosis Tool Using Component-Based SVM. In Proceedings of the 2008 IEEE Nuclear Science Symposium Conference Record, Dresden, Germany, 19–25 October 2008; pp. 4392–4395.
- Marcus, D.S.; Wang, T.H.; Parker, J.; Csernansky, J.G.; Morris, J.C.; Buckner, R.L. Open Access Series of Imaging Studies (OASIS): Cross-Sectional MRI Data in Young, Middle Aged, Nondemented, and Demented Older Adults. J. Cogn. Neurosci. 2007, 19, 1498–1507.
- Zhang, Y.; Dong, Z.; Phillips, P.; Wang, S.; Ji, G.; Yang, J.; Yuan, T.-F. Detection of Subjects and Brain Regions Related to Alzheimer’s Disease Using 3D MRI Scans Based on Eigenbrain and Machine Learning. Front. Comput. Neurosci. 2015, 9.
- Ding, Y.; Sohn, J.H.; Kawczynski, M.G.; Trivedi, H.; Harnish, R.; Jenkins, N.W.; Lituiev, D.; Copeland, T.P.; Aboian, M.S.; Mari Aparici, C.; et al. A Deep Learning Model to Predict a Diagnosis of Alzheimer Disease by Using 18F-FDG PET of the Brain. Radiology 2019, 290, 456–464.
- Rathore, S.; Habes, M.; Iftikhar, M.A.; Shacklett, A.; Davatzikos, C. A Review on Neuroimaging-Based Classification Studies and Associated Feature Extraction Methods for Alzheimer’s Disease and Its Prodromal Stages. Neuroimage 2017, 155, 530–548.
- Chang, C.-H.; Lin, C.-H.; Lane, H.-Y. Machine Learning and Novel Biomarkers for the Diagnosis of Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 2761.
- Stamate, D.; Kim, M.; Proitsi, P.; Westwood, S.; Baird, A.; Nevado-Holgado, A.; Hye, A.; Bos, I.; Vos, S.J.B.; Vandenberghe, R.; et al. A Metabolite-Based Machine Learning Approach to Diagnose Alzheimer-Type Dementia in Blood: Results from the European Medical Information Framework for Alzheimer Disease Biomarker Discovery Cohort. Alzheimer’s Dement. 2019, 5, 933–938.
- Binaco, R.; Calzaretto, N.; Epifano, J.; McGuire, S.; Umer, M.; Emrani, S.; Wasserman, V.; Libon, D.J.; Polikar, R. Machine Learning Analysis of Digital Clock Drawing Test Performance for Differential Classification of Mild Cognitive Impairment Subtypes Versus Alzheimer’s Disease. J. Int. Neuropsychol. Soc. 2020, 26, 690–700.
- Spasov, S.; Passamonti, L.; Duggento, A.; Liò, P.; Toschi, N. A Parameter-Efficient Deep Learning Approach to Predict Conversion from Mild Cognitive Impairment to Alzheimer’s Disease. NeuroImage 2019, 189, 276–287.
- Bae, J.; Stocks, J.; Heywood, A.; Jung, Y.; Jenkins, L.; Hill, V.; Katsaggelos, A.; Popuri, K.; Rosen, H.; Beg, M.F.; et al. Transfer Learning for Predicting Conversion from Mild Cognitive Impairment to Dementia of Alzheimer’s Type Based on a Three-Dimensional Convolutional Neural Network. Neurobiol. Aging 2020, 99, 53–64.
- Cabral, C.; Morgado, P.M.; Costa, D.C.; Silveira, M. Alzheimer’s Disease Neuroimaging Initiative Predicting Conversion from MCI to AD with FDG-PET Brain Images at Different Prodromal Stages. Comput. Biol. Med. 2015, 58, 101–109.
- Cheng, B.; Liu, M.; Zhang, D.; Munsell, B.C.; Shen, D. Domain Transfer Learning for MCI Conversion Prediction. IEEE Trans. Biomed. Eng. 2015, 62, 1805–1817.
- Cuingnet, R.; Gerardin, E.; Tessieras, J.; Auzias, G.; Lehéricy, S.; Habert, M.-O.; Chupin, M.; Benali, H.; Colliot, O. Alzheimer’s Disease Neuroimaging Initiative Automatic Classification of Patients with Alzheimer’s Disease from Structural MRI: A Comparison of Ten Methods Using the ADNI Database. Neuroimage 2011, 56, 766–781.
- Davatzikos, C.; Bhatt, P.; Shaw, L.M.; Batmanghelich, K.N.; Trojanowski, J.Q. Prediction of MCI to AD Conversion, via MRI, CSF Biomarkers, Pattern Classification. Neurobiol. Aging 2011, 32, 2322.e19–>2322.e27.
- Engedal, K.; Barca, M.L.; Høgh, P.; Bo Andersen, B.; Dombernowsky, N.W.; Naik, M.; Gudmundsson, T.E.; Øksengaard, A.-R.; Wahlund, L.-O.; Snaedal, J. The Power of EEG to Predict Conversion from Mild Cognitive Impairment and Subjective Cognitive Decline to Dementia. Dement. Geriatr. Cogn. Disord. 2020, 49, 38–47.
- Grassi, M.; Rouleaux, N.; Caldirola, D.; Loewenstein, D.; Schruers, K.; Perna, G.; Dumontier, M. Alzheimer’s Disease Neuroimaging Initiative a Novel Ensemble-Based Machine Learning Algorithm to Predict the Conversion from Mild Cognitive Impairment to Alzheimer’s Disease Using Socio-Demographic Characteristics, Clinical Information, and Neuropsychological Measures. Front. Neurol. 2019, 10, 756.
- Lee, G.; Nho, K.; Kang, B.; Sohn, K.-A.; Kim, D. Predicting Alzheimer’s Disease Progression Using Multi-Modal Deep Learning Approach. Sci. Rep. 2019, 9.
- Lin, W.; Gao, Q.; Yuan, J.; Chen, Z.; Feng, C.; Chen, W.; Du, M.; Tong, T. Predicting Alzheimer’s Disease Conversion from Mild Cognitive Impairment Using an Extreme Learning Machine-Based Grading Method with Multimodal Data. Front. Aging Neurosci. 2020, 12.
- Liu, S.; Liu, S.; Cai, W.; Che, H.; Pujol, S.; Kikinis, R.; Feng, D.; Fulham, M.J. ADNI Multimodal Neuroimaging Feature Learning for Multiclass Diagnosis of Alzheimer’s Disease. IEEE Trans. Biomed. Eng. 2015, 62, 1132–1140.
- Moradi, E.; Pepe, A.; Gaser, C.; Huttunen, H.; Tohka, J. Machine Learning Framework for Early MRI-Based Alzheimer’s Conversion Prediction in MCI Subjects. NeuroImage 2015, 104, 398–412.
- Pan, D.; Zeng, A.; Jia, L.; Huang, Y.; Frizzell, T.; Song, X. Early Detection of Alzheimer’s Disease Using Magnetic Resonance Imaging: A Novel Approach Combining Convolutional Neural Networks and Ensemble Learning. Front. Neurosci. 2020, 14, 259.
- Platero, C.; Tobar, M.C. Alzheimer’s Disease Neuroimaging Initiative Predicting Alzheimer’s Conversion in Mild Cognitive Impairment Patients Using Longitudinal Neuroimaging and Clinical Markers. Brain Imaging Behav. 2020.
- Popescu, S.G.; Whittington, A.; Gunn, R.N.; Matthews, P.M.; Glocker, B.; Sharp, D.J.; Cole, J.H. Alzheimer’s Disease Neuroimaging Initiative Nonlinear Biomarker Interactions in Conversion from Mild Cognitive Impairment to Alzheimer’s Disease. Hum. Brain Mapp. 2020.
- Pusil, S.; Dimitriadis, S.I.; López, M.E.; Pereda, E.; Maestú, F. Aberrant MEG Multi-Frequency Phase Temporal Synchronization Predicts Conversion from Mild Cognitive Impairment-to-Alzheimer’s Disease. Neuroimage Clin. 2019, 24, 101972.
- Tong, T.; Gao, Q.; Guerrero, R.; Ledig, C.; Chen, L.; Rueckert, D. A Novel Grading Biomarker for the Prediction of Conversion from Mild Cognitive Impairment to Alzheimer’s Disease. IEEE Trans. Biomed. Eng. 2017, 64, 155–165.
- Yan, Y.; Somer, E.; Grau, V. Classification of Amyloid PET Images Using Novel Features for Early Diagnosis of Alzheimer’s Disease and Mild Cognitive Impairment Conversion. Nucl. Med. Commun. 2019, 40, 242–248.
- Petersen, R.C. How Early Can We Diagnose Alzheimer Disease (and Is It Sufficient)? The 2017 Wartenberg Lecture. Neurology 2018, 91, 395–402.
- Grundman, M.; Petersen, R.C.; Ferris, S.H.; Thomas, R.G.; Aisen, P.S.; Bennett, D.A.; Foster, N.L.; Jack, C.R.; Galasko, D.R.; Doody, R.; et al. Mild Cognitive Impairment Can Be Distinguished from Alzheimer Disease and Normal Aging for Clinical Trials. Arch. Neurol. 2004, 61, 59–66.
- Kohannim, O.; Hua, X.; Hibar, D.P.; Lee, S.; Chou, Y.-Y.; Toga, A.W.; Jack, C.R., Jr.; Weiner, M.W.; Thompson, P.M. Boosting Power for Clinical Trials Using Classifiers Based on Multiple Biomarkers. Neurobiol. Aging 2010, 31, 1429–1442.
- Walhovd, K.B.; Fjell, A.M.; Brewer, J.; McEvoy, L.K.; Fennema-Notestine, C.; Hagler, D.J.; Jennings, R.G.; Karow, D.; Dale, A.M. Combining MR Imaging, Positron-Emission Tomography, and CSF Biomarkers in the Diagnosis and Prognosis of Alzheimer Disease. Am. J. Neuroradiol. 2010, 31, 347–354.
- Westman, E.; Muehlboeck, J.-S.; Simmons, A. Combining MRI and CSF Measures for Classification of Alzheimer’s Disease and Prediction of Mild Cognitive Impairment Conversion. Neuroimage 2012, 62, 229–238.
- Gao, J.; Li, P.; Chen, Z.; Zhang, J. A Survey on Deep Learning for Multimodal Data Fusion. Neural Comput. 2020, 32, 829–864.
- El-Sappagh, S.; Alonso, J.M.; Islam, S.M.R.; Sultan, A.M.; Kwak, K.S. A Multilayer Multimodal Detection and Prediction Model Based on Explainable Artificial Intelligence for Alzheimer’s Disease. Sci. Rep. 2021, 11, 2660.
- Sancesario, G.M.; Toniolo, S.; Chiasserini, D.; Di Santo, S.G.; Zegeer, J.; Bernardi, G.; Musicco, M.; Caltagirone, C.; Parnetti, L.; Bernardini, S. The Clinical Use of Cerebrospinal Fluid Biomarkers for Alzheimer’s Disease Diagnosis: The Italian Selfie. J. Alzheimer’s Dis. 2017, 55, 1659–1666.
- Mattsson, N.; Palmqvist, S.; Stomrud, E.; Vogel, J.; Hansson, O. Staging β-Amyloid Pathology with Amyloid Positron Emission Tomography. JAMA Neurol. 2019, 76, 1319–1329.
- Rabinovici, G.D.; Gatsonis, C.; Apgar, C.; Chaudhary, K.; Gareen, I.; Hanna, L.; Hendrix, J.; Hillner, B.E.; Olson, C.; Lesman-Segev, O.H.; et al. Association of Amyloid Positron Emission Tomography with Subsequent Change in Clinical Management Among Medicare Beneficiaries with Mild Cognitive Impairment or Dementia. JAMA 2019, 321, 1286–1294.
- Ossenkoppele, R.; Rabinovici, G.D.; Smith, R.; Cho, H.; Schöll, M.; Strandberg, O.; Palmqvist, S.; Mattsson, N.; Janelidze, S.; Santillo, A.; et al. Discriminative Accuracy of [18F]Flortaucipir Positron Emission Tomography for Alzheimer Disease vs. Other Neurodegenerative Disorders. JAMA 2018, 320, 1151–1162.
- Preische, O.; Schultz, S.A.; Apel, A.; Kuhle, J.; Kaeser, S.A.; Barro, C.; Gräber, S.; Kuder-Buletta, E.; LaFougere, C.; Laske, C.; et al. Serum Neurofilament Dynamics Predicts Neurodegeneration and Clinical Progression in Presymptomatic Alzheimer’s Disease. Nat. Med. 2019, 25, 277–283.
- Palmqvist, S.; Janelidze, S.; Stomrud, E.; Zetterberg, H.; Karl, J.; Zink, K.; Bittner, T.; Mattsson, N.; Eichenlaub, U.; Blennow, K.; et al. Performance of Fully Automated Plasma Assays as Screening Tests for Alzheimer Disease-Related β-Amyloid Status. JAMA Neurol. 2019, 76, 1060–1069.
- Schindler, S.E.; Bollinger, J.G.; Ovod, V.; Mawuenyega, K.G.; Li, Y.; Gordon, B.A.; Holtzman, D.M.; Morris, J.C.; Benzinger, T.L.S.; Xiong, C.; et al. High-Precision Plasma β-Amyloid 42/40 Predicts Current and Future Brain Amyloidosis. Neurology 2019, 93, e1647–e1659.
- Janelidze, S.; Mattsson, N.; Palmqvist, S.; Smith, R.; Beach, T.G.; Serrano, G.E.; Chai, X.; Proctor, N.K.; Eichenlaub, U.; Zetterberg, H.; et al. Plasma P-Tau181 in Alzheimer’s Disease: Relationship to Other Biomarkers, Differential Diagnosis, Neuropathology and Longitudinal Progression to Alzheimer’s Dementia. Nat. Med. 2020, 26, 379–386.
- Palmqvist, S.; Tideman, P.; Cullen, N.; Zetterberg, H.; Blennow, K.; Dage, J.L.; Stomrud, E.; Janelidze, S.; Mattsson-Carlgren, N.; Hansson, O. Prediction of Future Alzheimer’s Disease Dementia Using Plasma Phospho-Tau Combined with Other Accessible Measures. Nat. Med. 2021.
More
Information
Subjects:
Neurosciences
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Revisions:
2 times
(View History)
Update Date:
03 Sep 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




