| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Patrick Romano Monteiro | + 3125 word(s) | 3125 | 2021-08-06 10:59:03 | | | |
| 2 | Amina Yu | Meta information modification | 3125 | 2021-08-16 03:21:10 | | |
Video Upload Options
Anabaenopeptins (APs) are structurally diverse peptides widely distributed in distinct ecosystems among cyanobacteria. Some structural features of these molecules are shared with other cyanotoxins, such as the presence of modified residues, exocyclic amino acids, circular structure, and amino acids in D-configuration. However, among the cyanopeptides, the ureido linkage is exclusively found in APs. Thus, these cyclic peptides demonstrate toxicity and structural diversity which will be explored in this topic, including biotechnological and ecological relevance, and their distribution.
1. Introduction
Cyanobacteria are photosynthetic microorganisms widely distributed in the world. They can inhabit several types of ecosystems, including aquatic and terrestrial. These microorganisms produce a great variety of bioactive compounds, which have been investigated mainly due to their biotechnological potential and environmental relevance [1][2][3]. Cyanotoxins are among the most studied compounds originated from cyanobacteria since they are capable of negatively affecting human and animal health [4][5]. These metabolites can vary drastically concerning their action mechanism and chemical structure, which include peptides, alkaloids, and lipopolysaccharides [6][7][8]. The majority of publications related to peptides from cyanobacteria have mainly focused on the class of microcystins with over 300 characterized variants [9][10]. However, cyanobacteria usually do not exclusively produce a single class of compounds, given that specific strains are co-producing different groups of secondary metabolites [11].
Other peptides beyond microcystins have been poorly explored, lacking information mainly in the environmental sciences [11]. These several metabolites are known for their potent inhibitory properties against several enzymes in nanomolar concentrations, resulting in toxic effects [12][13]. Moreover, similar to microcystins, they have been regularly detected in diverse environments [14]. In certain regions, their occurrence is more pronounceable than microcystins themselves [15]. However, information about the concentrations which are encountered is rarely reported [11]. Cyanobacteria have developed different peptides as a protection mechanism against parasites [16]. Concerning their origin, some peptides as microviridins and cyanobactins are produced via ribosomal whereas others as microginins and aeruginosins are synthesized by non-ribosomal pathways [13][17][18].
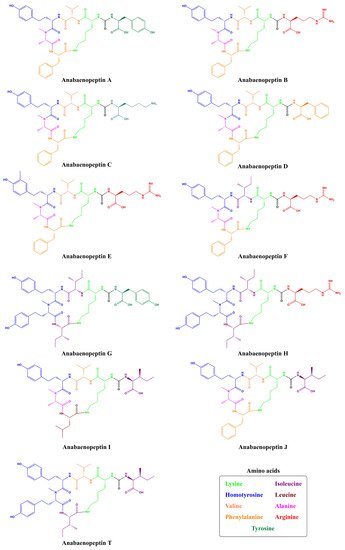
Among the most recurrent peptides encountered in the environment are anabaenopeptins (APs), a family of cyclic peptides containing six amino acid residues [19]. They have been found in an enormous variety of cyanobacteria isolated from both the aquatic and terrestrial environments, including Anabaena , Nostoc , Microcystis , Planktothrix , Lyngbya, and Brasilonema [12][20][21][22][23][24]. In their general structure is a well-conserved Lysine (Lys) residue in D-configuration, which is responsible for the ring formation and five additional variable amino acids, either proteinogenic or non-proteinogenic, resulting in 124 described AP variants from cyanobacteria (Supplementary Table 1) [19]. Besides their structural variety, molecules belonged to this group exhibit an impressive functional diversity, which includes inhibitory activity for proteases, phosphatases, and carboxypeptidases [22][25][26].
The enormous structural diversification of anabaenopeptins can be attributed to the low substrate specificity of some enzymes involved in their synthesis as well as the presence of alternative starter modules [16]. Their production is strongly influenced by environmental factors [27]. Besides that, because of their diversified bioactive properties, they exhibit an elevated biotechnological potential. This review aims at presenting the main researches on anabaenopeptins, emphasizing their general characteristics, biosynthesis as well as ecological and biotechnological relevance.
2. Structures of Anabaenopeptins
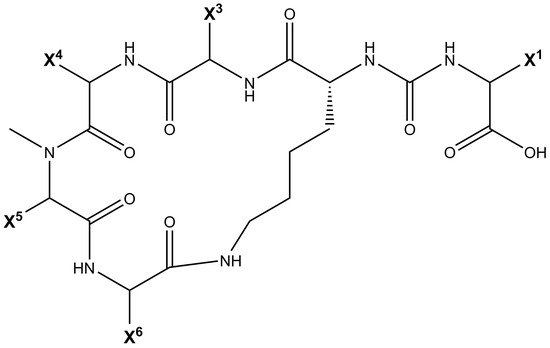
Being non-ribosomally synthesized, anabaenopeptin structures comprise a ring of five amino acids connected through an ureido linkage to an exocyclic amino acid. Thus, its general structure is represented by X 1-CO-[Lys 2-X 3-X 4-MeX 5-X 6], where the bracket represents the cyclic region of this peptide and X are variable amino acids according to their positions represented by the superscript numbers ( Figure 1 ). Its ring is formed by cyclization of the C-terminal carboxyl of the amino acid at position 6 to the ε-NH 3 of the well-conserved D-lysine at position 2. Furthermore, the α-amino group of Lys is connected to the exocyclic amino acid X 1 via an ureido bridge. Due to its non-ribosomal nature, proteinogenic and non-proteinogenic amino acids are usually detected in this hexapeptide [19].
Some of those conserved features from APs can also be visualized in other cyanopeptides. Veraguamides A-G are cyclic hexadepsipeptides, and they do not possess any exocyclic residue. Lyngbyastatin peptides demonstrate elastase, trypsin, and chymotrypsin inhibitory properties. Their structures consist of a 6-member ring coupled to a chain of 2 exocyclic residues and can bear modified and unusual residues. Also possessing a 6-member ring structure and 2 exocyclic amino acids, Tiglicamides A-C were obtained from Lyngbya confervoides [28].
Cyanopeptolins are depsipeptides containing a 6-amino acid ring bearing a side chain with 1–2 residues and modified residues, such as 3-amino-6-hydroxy-2-piperidone. Cyanopeptolin A is one example of this class of cyanopeptides and is composed by (1)-fatty acid, (2)-Arg, (3)-Ahp, (4)-Leu, (5)-methyl-Phe, (6)-Val, and (7)-Thr, in this case, the β-lacton ring is formed between Arg and Thr residues and positions 2, 4, 5 and 6 are variable. Using Anabaenopeptin A as reference ( Figure 2 ), its structure is (1)-Tyr, (2)-D-Lys, (3)-Valine, (4)-Homotyrosine, (5)N-methyl-Alanine, (6)-Phenylalanine [29]. Positions 1, 3, 4, 5, and 6 are variable concerning APs ( Figure 1 ) and the ureido bond is formed between 1 and 2 residues. Aerucyclamides are entirely cyclic peptides, Aerucyclamide A is composed by (1)-dehydro-Thr, (2)-Gly, (3)-thiozole, (4)-Ile, (5)-dhCys, and (6)-Ile, in this case, variations were reported in positions 2, 3, 4 and 6. Different from the cyanopeptides listed until now, Aeruginosins and Microginins are linear peptides. Aeruginosin KB 676 is formed by (1)-Hpla, (2)-Ile, (3)-Choi and (4)-Arg, only position 2 presents variation with amino acid substitution, and radical changes occur in positions 3 and 4. Finally, Microginin 713 is formed by (1)-Ahda, (2)-Ala, (3)-Val, (4)-N-methyl-Tyr, and (5)-Tyr, in this case, positions 2, 3, 4, and 5 had substitutions reported [11].
Structurally, despite the major amino acid variability, Microcystins, Cyanopeptolins, and Anabaenopeptins are most similar. Microcystins and Cyanopeptolins are heptapeptides and Anabaenopeptins are hexapeptides and comparting these structures, it is possible to distinguish a ring core and a linear region. Although Microcystins are technically cyclic peptides, the Adda moiety projected outside the ring may act like the fatty acid in Cyanopeptolin A or Tyrosine in Anabaenopeptin A. The Adda moiety is crucial for MCs inhibition towards phosphatases, as its long linear chain can penetrate the enzyme active site together with other side chains, having a similar role as the exocyclic residue of APs (Tyrosine from Anabaenopeptin A), as it will be further discussed in Section 7 [11][34]. This exocyclic or even protuberant residue was not observed in Aerucyclamides that only present a cyclic structure or Aeruginosins and Microginins, which are linear structures [11]. Therefore, cyclic peptides bearing exocyclic residues and unusual and D-configuration amino acids are also found in cyanobacteria, however, the ureido linkage in cyanopeptides is, so far, an exclusive characteristic of Anabaenopeptins.
3. Occurrence of Anabaenopeptins and factors involved in their expression
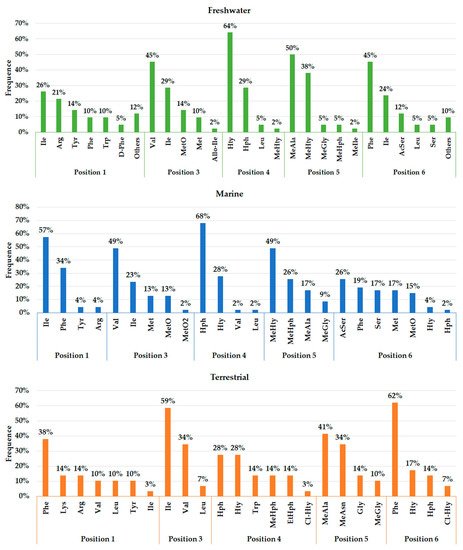
A total of 45, 29, and 12 cyanobacteria strains from freshwater, marine and terrestrial environment have been analyzed for AP production, respectively. As seen in Table 1 and according to the literature [35][36][33][37][38][39][40][41][42][43][44][45][46][47], marine strains produced a total of 50 different variants of APs, in comparison to 43 and 34 variants from freshwater, and terrestrial strains, respectively ( Figure 3 ). Thus, marine cyanobacteria demonstrate to produce a higher number of distinct APs variants in comparison to the remaining strains from different sources. However, APs from freshwater environments have the greatest diversity of amino acids in the majority of positions ( Figure 4 ). Thus, these features could be associated with different obstacles faced in their respective environments as well as the fact that both belong to aquatic environments [48], however, this hypothesis requires further studies. Some of those APs are shared among different strains isolated from distinct environments: 2 anabaenopeptins (A and B) variants were detected in all ecosystems; in comparison, strains from both aquatic habitats had 13 APs variants in common (D, F, J, 807, NZ841, Oscillamide Y, and Nodulapeptins B, C, 855B, 871, 879, 897 and 915A) ; in contrast, only anabaenopeptin C were produced by both terrestrial and freshwater, and none Anabaenopeptin variant was shared by both terrestrial and marine strains.
| Strains | Anabaenopeptin | Reference |
|---|---|---|
| Freshwater | ||
| Anabaena flos-aquae 202 A 1 | Anabaenopeptins B and D | [29] |
| Anabaena flos-aquae CYA 83/1 | Anabaenopeptins B and D | [29] |
| Anabaena lemmermannii 202 A2/41 | Anabaenopeptins B and C | [29] |
| Aphanizomenon flos-aquae NIES-81 | Anabaenopeptins I and J | [32] |
| Lyngbya sp. (SAG 36.91) | Lyngbyaureamide A and B | [49] |
| Microcystis aeruginosa HUB 063 | Anabaenopeptins B and F | [36] |
| Microcystis aeruginosa Kutz | Ferintoic acids A and B | [50] |
| Microcystis aeruginosa PCC7806 | Anabaenopeptins A, B, E/F and Oscillamide Y | [51] |
| Microcystis aeruginosa TAU IL-342 | Anabaenopeptin HU892 | [52] |
| Microcystis sp. (MB-K) | Anabaenopeptin KT864 | [53] |
| Microcystis sp. TAU IL-306 | Anabaenopeptin F and Oscillamide Y | [52] |
| Microcystis sp. TAU IL-362 | Anabaenopeptins MM823, MM850, MM913 and B | [52] |
| Microcystis spp. | Anabaenopeptin KB905, KB899, G, H, 908A, 915, HU892, MM913 | [54] |
| Nodularia spumigena Node 2 | Nodulapeptins B, C, 855B, 871, 879, 897 and 915A | [14][55] |
| Nodularia spumigena Nodg 3 | Nodulapeptins B, C, 855B, 871, 879, 897 and 915A | [14] |
| Nodularia spumigena Nodh 2 | Nodulapeptins B, C, 855B, 871, 879, 897 and 915A | [14] |
| Nodularia spumigena NSBL-05 | Anabaenopeptin 807 | [14] |
| Nodularia spumigena NSBL-06 | Anabaenopeptin 807 | [14] |
| Nodularia spumigena NSBR-01 | Anabaenopeptin 807 | [14] |
| Nodularia spumigena NSGL-01 | Anabaenopeptin 807 | [14] |
| Nodularia spumigena NSKR-07 | Anabaenopeptin 807 | [14] |
| Nodularia spumigena NSLA-01 | Anabaenopeptin 807 | [14] |
| Nodularia spumigena NSOR-02 | Anabaenopeptin 807 | [14] |
| Nodularia spumigena NSPH-02 | Anabaenopeptin 807 | [14] |
| Oscillatoria agardhii CYA 128 | Anabaenopeptins A and C | [29] |
| Oscillatoria agardhii NIES-204 | Anabaenopeptins B, E and F | [31] |
| Oscillatoria agardhii NIES-595 | Anabaenopeptin G and H | [26] |
| Planktothrix agardhii CCAP 1459/11A | Anabaenopeptin F and Oscillamide B | [25] |
| Planktothrix agardhii CYA126/8 | Anabaenopeptin 908A and 915 | [56] |
| Planktothrix agardhii HUB 011 | Anabaenopeptin G | [36] |
| Planktothrix agardhii NIVA CYA 15 | Anabaenopeptins A and B | [57] |
| Planktothrix agardhii NIVA CYA 34 | Anabaenopeptins A, B, F and Oscillamide Y | [57] |
| Planktothrix mougeotii NIVA CYA 405 | Anabaenopeptins A, B, F and Oscillamide Y | [57] |
| Planktothrix mougeotii NIVA CYA 56/3 | Anabaenopeptins C, 822 *, B, and F | [57] |
| Planktothrix prolifica NIVA CYA 406 | Anabaenopeptins A, B, F and Oscillamide Y | [57] |
| Planktothrix prolifica NIVA CYA 540 | Anabaenopeptins A, B, F and Oscillamide Y | [57] |
| Planktothrix prolifica NIVA CYA 98 | Anabaenopeptins A, B, F and Oscillamide Y | [18][57] |
| Planktothrix rubescens | Anabaenopeptins A, B, F and Oscillamide Y | [58] |
| Planktothrix rubescens | Anabaenopeptins A, B, C, F and Oscillamide Y | [59] |
| Planktothrix rubescens | Anabaenopeptins B and F | [60] |
| Planktothrix rubescens | Anabaenopeptin A, B, and F | [61] |
| Planktothrix rubescens BGSD-500 | Anabaenopeptins B and F | [62] |
| Planktothrix rubescens NIES-610 | Anabaenopeptin F | [25] |
| Planktothrix rubescens NIVA CYA 407 | Anabaenopeptins C, 822 *, B, and F | [57] |
| Woronichinia naegeliana | Anabaenopeptin 899 | [63] |
| Marine | ||
| Anabaena sp. TAU NZ-3-1 | Anabaenopeptins NZ841, NZ825 and NZ857 | [64] |
| Coelosphaeriaceae cyanobacterium 06S067 | Anabaenopeptins A, B, F, 802 *, 827 *, 809 * and Oscillamide Y | [65] |
| Nodularia spumigena AV1 | Nodulapeptins A, B, C, 871, 821, 839, 849, 855A, 863, 865, 867, 879, 881A, 881B, 883A, 897, 899A, 915A, 931 | [14][66][55] |
| Nodularia spumigena B15a | Anabaenopeptins 841 and D | [14] |
| Nodularia spumigena BY1 | Anabaenopeptin B and Nodulapeptins B, C, 821, 839, 855A, 855B, 871, 879, 881A, 881B, 883A, 897, 899A, 915A, 931 | [14][66][55] |
| Nodularia spumigena CCNP 1401 | Anabaenopeptins 841A and D | [14][55] |
| Nodularia spumigena CCNP 1423 | Nodulapeptins 883B, 899B, 901, 915B, 917, 933 | [14][55] |
| Nodularia spumigena CCNP 1424 | Nodulapeptins 883B, 899B, 901, 915B, 917, 933 | [14][55] |
| Nodularia spumigena CCNP 1425 | Nodulapeptins 883B, 899B, 901, 915B, 917, 933 | [14][55] |
| Nodularia spumigena CCNP 1402 | and Nodulapeptins A, B, C, 821, 839, 855A, 855B, 871, 879, 881A, 881B, 883A, 897, 899A, 915A, 931 | [14][55] |
| Nodularia spumigena CCNP 1403 | Anabaenopeptins 841A and D | [14][55] |
| Nodularia spumigena CCNP 1426 | Anabaenopeptins D and 841A | [55] |
| Nodularia spumigena CCNP 1427 | Nodulapeptins B, C, 821, 855A, 855B, 871, 879, 881A, 881B, 883A, 897, 899A, 915A and 931 | [55] |
| Nodularia spumigena CCNP 1428 | Nodulapeptins 883B, 899B, 901, 915B, 917 and 933 | [55] |
| Nodularia spumigena CCNP 1430 | Anabaenopeptins D and 841A | [55] |
| Nodularia spumigena CCNP 1431 | Nodulapeptins 883B, 885, 899B, 901, 915B, 917 and 933 | [55] |
| Nodularia spumigena CCNP 1436 | Nodulapeptins B, C, 839, 855A, 855B, 871, 879, 881A, 881B, 883A, 897, 899A, 915A, 921 and 931 | [55] |
| Nodularia spumigena CCNP 1440 | Nodulapeptins 883B, 885, 899B, 901, 915B, 917 and 933 | [55] |
| Nodularia spumigena CCY 9414 | Nodulapeptins A, B, C, 839, 855A, 855B, 871, 879, 881A, 881B, 883A, 897, 899A, 915A, 931 | [14][55][67] |
| Nodularia spumigena KAC 11 | Anabaenopeptins J and 807 | [55] |
| Nodularia spumigena KAC 13 | Anabaenopeptins D and 841A | [55] |
| Nodularia spumigena KAC 64 | Nodulapeptins 883B, 885, 899B, 901, 915B, 917 and 933 | [55] |
| Nodularia spumigena KAC 66 | Nodulapeptins 883B, 885, 857, 899B, 901, 915B, 917 and 933 | [14][55] |
| Nodularia spumigena KAC 68 | Nodulapeptins 883B, 885, 857, 899B, 901, 917 and 933 | [55] |
| Nodularia spumigena KAC 7 | Nodulapeptins B, C, 921, 839, 855A, 855B, 871, 879, 881A, 881B, 883A, 897, 899A, 915A and 931 | [55] |
| Nodularia spumigena KAC 70 | Nodulapeptins 807, 823, 851, 865, 867 and 883C | [55] |
| Nodularia spumigena KAC 71 | Nodulapeptins A, B, C, 921, 823, 839, 855A, 855B, 871, 879, 881A, 881B, 883A, 897, 899A, 915A and 931 | [55] |
| Nodularia spumigena KAC 87 | Nodulapeptins 807, 823, 849, 851, 865, 867 and 883C | [55] |
| Nodularia spumigena UHCC0039 | Nodulapeptins A, B, C, 839, 849, 855A, 863, 865, 867, 871, 879, 881A, 881B, 897, 899A, 915A and 933 | [67] |
| Terrestrial | ||
| Anabaena circinalis 90 | Anabaenopeptins A, B, and C | [29] |
| Anabaena flos-aquae NRC 525-17 | Anabaenopeptins A and B | [20] |
| Brasilonema sp. 360 | Anabaenopeptin 802A | [24] |
| Brasilonema sp. 382 | Anabaenopeptin 802A | [24] |
| Brasilonema sp. CT11 | Anabaenopeptins 788, 802A, 802B and 816 | [68] |
| Desmonostoc sp. 386 | Anabaenopeptins 848, 849, 862, 863, 877A, 877B, 891 and 905 | [24] |
| Nostoc sp. 352 | Anabaenopeptins 841B, 855, 857 and 871 | [24] |
| Nostoc sp. 358 | Anabaenopeptins 882 and 896 | [24] |
| Nostoc sp. ASN_M | Anabaenopeptins 808 *, 828, 842 *, 844 * and 858 *, | [69] |
| Nostoc sp. ATCC 53789 | Anabaenopeptin SA9, SA10, SA11 and SA12 | [12] |
| Nostoc sp. KVJ2 | Anabaenopeptins KVJ827, KVJ841, and KVJ811 | [21] |
| Schizothrix sp. IL-208-2-2 | Schizopeptin 791 | [70] |
* Anabaenopeptin variants with non-elucidated sequence.
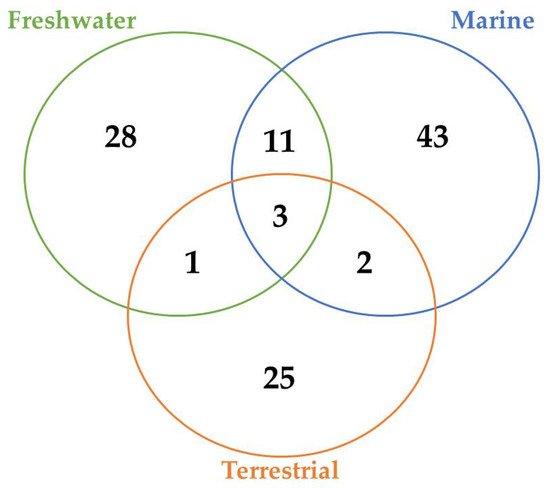
According to Table 1 and Figure 3 , there are AP variants shared among cyanobacteria strains from different environments according to the previous discussion. Anabaenopeptins A and B are the only variants detected in all habitats analyzed, and the only difference between those variants resides at the exocyclic residue. AP B is still the most recurrent among these oligopeptides in cyanobacteria ( Table 1 ), corroborating with the previously raised hypothesis that this variant was the first cyanotoxin of this class to be emerged. [71]. Furthermore, the number of common anabaenopeptins variants increases when a comparison is made among strains only from aquatic habitats (freshwater and marine): Anabaenopeptins D, F, J, 807, NZ841, Oscillamide Y, and Nodulapeptins B, C, 855B, 871, 879, 897 and 915A. Besides their production by both freshwater and marine cyanobacteria, these prevalent oligopeptides seem to be more recurrent in marine environments, given that a higher number of cyanobacteria strains from this habitat are able to produce these APs comparing to freshwater, except for Oscillamide Y, which is more recurring in the latter. Among those variants, Nodulapeptin B is the most frequent in marine microorganisms. Besides, the only difference between the AP C (produced by freshwater and terrestrial strains) and both A and B variants is the exocyclic amino acid, and the former was not detected in marine cyanobacteria.
As seen in Figure 3 , the environment can exert a crucial role in the biosynthesis of different APs, justifying their distribution in certain locations. The presence and frequency of certain amino acids in Anabaenopeptin structures can vary according to their respective source environment. Anabaenopeptins from both aquatic environments demonstrate to have Isoleucine as the most recurrent amino acid in position 1, while this same amino acid was detected in only one AP variant in terrestrial strains ( Figure 4 ). Phenylalanine was highly detected in position 1 of Anabaenopeptins isolated from terrestrial strains. Then, freshwater cyanobacteria may be promising biotechnological targets due to its highest diversity of amino acids in position 1, as the exocyclic residue is crucial for its inhibitory activity [12][35][34]. Regarding the variable position 3, Anabaenopeptins from freshwater and marine environments displayed a similar pattern of amino acid frequencies, Valine (Val) being the most frequent, followed by Ile and L-Methionine sulfone (MetO2). In contrast, terrestrial strains produce several AP variants with Ile in position 3, followed by Val and Leu, the latter being absent in this position on APs detected in aquatic environments. Homotyrosine (Hty) and Homophenylalanine (Hph) are the most found residues in position 4 among APs from all habitats analyzed, however, among terrestrial and marine strains Hph is more predominantly, while Hty is commonly observed in APs from freshwater strains. Except for Glycine (Gly) in some Anabaenopeptins from terrestrial strains, all the other residues in position 5 are N-methylated. APs from non-aquatic cyanobacteria do not harbor homoamino acids in the fifth position and, in addition, Asparagine is only detected in some of those variants in the respective position. Besides their detection in position 5, homoamino acids seem to be more persistent in position 4 from those APs analyzed. Position 6 has the highest richness of amino acids among AP variants obtained from marine environments, having incorporated 7 different residues, while this position in variants from freshwater habitats have assimilated 9 different amino acids, being the second most diverse site. Such heterogeneity in the last position in APs from aquatic strains is not clear, as the first amino acid residue demonstrated to be important in Anabaenopeptin interaction towards its enzyme target [12][35][34]. This array of several amino acids detected in position 6 is not visualized in Anabaenopeptins from terrestrial strains, where Phe was the amino acid more detected, similar to those APs from freshwater microorganisms.
In addition to interaction with other cyanobacteria, these microorganisms are capable to establish symbiotic associations with invertebrates, such as corals, mollusks, and sponges. Both organisms can be benefited during this consortium through secondary metabolite production, for example [72]. Sponges host an enormous quantity of microorganisms belonging to diverse phyla, where cyanobacteria are mainly represented by genera Aphanocapsa , Synechocystis , Phormidium , and Oscillatoria [73]. These photosynthetic microorganisms can occupy either extra- or intracellular spaces, aiding the host in the control of the redox potential, supplying pigments and energy through carbon fixation, and in the defense mechanism by the production of secondary metabolites. Published reports have demonstrated that as a consequence of these processes, cyanobacteria have their metabolic profile altered, resulting in the production of distinct variants of natural products. The compound 2-(2’,4’-dibromophenyl)-4,6-dibromophenol is solely biosynthesized by a cyanobacterium belonging to genus Oscillatoria in association with the sponge Dysidea herbacea [74]. These factors corroborate with the hypothesis that anabaenopeptins primarily observed in sponges could be of cyanobacterial origin, as brominated APs variants were isolated only from sponges [75][76][77] and the Oscillatoria genus is known for APs production. For instance, the polyketide nosperin and some variants of oligopeptide nostopeptolide are encountered exclusively during symbiosis, which may be the same mechanism for anabaenopeptin variants production found in sponges.
4. Applications of Anabaenopeptins
Cyanopeptides such as APs have a well-demonstrated capacity of protease inhibition [78]. Protein Phosphatase 1 (PP1), Protein Phosphatase 2A, Carboxypeptidase-A (CPA), Human Serine Protease, Leucine Aminopeptidase, Trypsin, and Thrombin have already been tested against several cyanobacterial extracts and confirmed the catalysis blockage [11].
Serine/threonine protein phosphatases inhibition was also reported [22][25]. Nevertheless, several other cyanopeptides presented more effective IC 50 levels against elastase, such as some variants of lyngbyastatins, symplostatins, microvirins, and others. Concerning PP1, MCs remain the best inhibitor among all cyanopeptides [11]. IC 50 reported values to MCs and nodularins are from 1.1 to 1.9 nM as PP1 inhibitors [79]. In this case, APs remain promising candidates in Carboxypeptidase inhibition.
Cyanopeptides blooms events may present the production of different classes of cyanopeptides like MCs, APs, and cyanopeptolins. A few studies quantified cyanopeptides beyond Microcystins, even so, in 10 eutrophic lakes in the United States and Europe the cyanopeptides concentration including these 3 types of cyanopeptides were from <4 µg/L to >40 µg /L [11]. In wet weight, 2.1 mg of AP and 7.4 mg of Microcystin-LR were obtained from 1.7 kg of biomass in a water bloom of lake Teganuma (Japan) [33].
Besides some cyanopeptides presented anticancer activity, APs have been presented poor results in cytotoxic tests [80]. Anabaenopeptin B had been tested about its anticancer potential and did not demonstrate cytotoxic effects against N2a, MCF−7, and GH4 cells even at the 500 µg/mL concentration [81]. Despite anticancer activity was detected in Aliinostoc sp. CENA543 extract containing AP , it was not possible to attribute this effect exclusively to this class of oligopeptides because there were other cyanopeptides in the extract, and the exact AP was not identified [82]. No cytotoxic activity was presented by Nodulapeptins 883C, 869, 867, 865, and Anabaenopeptin 813 as well [35].
References
- do Amaral, S.C.; Santos, A.V.; da Cruz Schneider, M.P.; da Silva, J.K.R.; Xavier, L.P. Determination of Volatile Organic Compounds and Antibacterial Activity of the Amazonian Cyanobacterium Synechococcus sp. Strain GFB01. Molecules 2020, 25, 4744.
- Gradíssimo, D.G.; Xavier, L.P.; Santos, A.V. Cyanobacterial Polyhydroxyalkanoates: A Sustainable Alternative in Circular Economy. Molecules 2020, 25, 4331.
- de Oliveira, D.T.; da Costa, A.A.F.; Costa, F.F.; da Rocha Filho, G.N.; do Nascimento, L.A.S. Advances in the Biotechnological Potential of Brazilian Marine Microalgae and Cyanobacteria. Molecules 2020, 25, 2908.
- Huisman, J.; Codd, G.A.; Paerl, H.W.; Ibelings, B.W.; Verspagen, J.M.H.; Visser, P.M. Cyanobacterial blooms. Nat. Rev. Microbiol. 2018, 16, 471–483.
- Bittner, M.; Štern, A.; Smutná, M.; Hilscherová, K.; Žegura, B. Cytotoxic and Genotoxic Effects of Cyanobacterial and Algal Extracts—Microcystin and Retinoic Acid Content. Toxins 2021, 13, 107.
- Chen, G.; Wang, L.; Li, W.; Zhang, Q.; Hu, T. Nodularin induced oxidative stress contributes to developmental toxicity in zebrafish embryos. Ecotoxicol. Environ. Saf. 2020, 194, 110444.
- Zhu, H.; Sonoyama, T.; Yamada, M.; Gao, W.; Tatsuno, R.; Takatani, T.; Arakawa, O. Co-Occurrence of Tetrodotoxin and Saxitoxins and Their Intra-Body Distribution in the Pufferfish Canthigaster valentini. Toxins 2020, 12, 436.
- Moosová, Z.; Šindlerová, L.; Ambrůzová, B.; Ambrožová, G.; Vašíček, O.; Velki, M.; Babica, P.; Kubala, L. Lipopolysaccharides from Microcystis Cyanobacteria-Dominated Water Bloom and from Laboratory Cultures Trigger Human Immune Innate Response. Toxins 2019, 11, 218.
- Massey, I.Y.; Yang, F. A Mini Review on Microcystins and Bacterial Degradation. Toxins 2020, 12, 268.
- Jones, M.R.; Pinto, E.; Torres, M.A.; Dörr, F.; Mazur-Marzec, H.; Szubert, K.; Tartaglione, L.; Dell’Aversano, C.; Miles, C.O.; Beach, D.G.; et al. CyanoMetDB, a comprehensive public database of secondary metabolites from cyanobacteria. Water Res. 2021, 196, 117017.
- Janssen, E.M.L. Cyanobacterial peptides beyond microcystins—A review on co-occurrence, toxicity, and challenges for risk assessment. Water Res. 2019, 151, 488–499.
- Schreuder, H.; Liesum, A.; Lönze, P.; Stump, H.; Hoffmann, H.; Schiell, M.; Kurz, M.; Toti, L.; Bauer, A.; Kallus, C.; et al. Isolation, Co-Crystallization and Structure-Based Characterization of Anabaenopeptins as Highly Potent Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa). Sci. Rep. 2016, 6, 32958.
- do Amaral, S.C.; Monteiro, P.R.; da Neto, J.S.P.; Serra, G.M.; Gonçalves, E.C.; Xavier, L.P.; Santos, A.V. Current Knowledge on Microviridin from Cyanobacteria. Mar. Drugs 2021, 19, 17.
- Mazur-Marzec, H.; Kaczkowska, M.; Blaszczyk, A.; Akcaalan, R.; Spoof, L.; Meriluoto, J. Diversity of Peptides Produced by Nodularia spumigena from Various Geographical Regions. Mar. Drugs 2013, 11, 1–19.
- Gkelis, S.; Lanaras, T.; Sivonen, K. Cyanobacterial Toxic and Bioactive Peptides in Freshwater Bodies of Greece: Concentrations, Occurrence Patterns, and Implications for Human Health. Mar. Drugs 2015, 13, 6319–6335.
- Rohrlack, T.; Christiansen, G.; Kurmayer, R. Putative Antiparasite Defensive System Involving Ribosomal and Nonribosomal Oligopeptides in Cyanobacteria of the Genus Planktothrix. Appl. Environ. Microbiol. 2013, 79, 2642–2647.
- Martins, J.; Vasconcelos, V. Cyanobactins from Cyanobacteria: Current Genetic and Chemical State of Knowledge. Mar. Drugs 2015, 13, 6910–6946.
- Rounge, T.B.; Rohrlack, T.; Nederbragt, A.J.; Kristensen, T.; Jakobsen, K.S. A genome-wide analysis of nonribosomal peptide synthetase gene clusters and their peptides in a Planktothrix rubescens strain. BMC Genom. 2009, 10, 396.
- Welker, M.; Von Döhren, H. Cyanobacterial peptides—Nature’s own combinatorial biosynthesis. FEMS Microbiol. Rev. 2006, 30, 530–563.
- Harada, K.I.; Fujii, K.; Shimada, T.; Suzuki, M.; Sano, H.; Adachi, K.; Carmichael, W.W. Two cyclic peptides, anabaenopeptins, a third group of bioactive compounds from the cyanobacteriumAnabaena flos-aquae NRC 525-17. Tetrahedron Lett. 1995, 36, 1511–1514.
- Guljamow, A.; Kreische, M.; Ishida, K.; Liaimer, A.; Altermark, B.; Bähr, L.; Hertweck, C.; Ehwald, R.; Dittmann, E. High-Density Cultivation of Terrestrial Nostoc Strains Leads to Reprogramming of Secondary Metabolome. Appl. Environ. Microbiol. 2017, 83, 1–15.
- Zafrir-Ilan, E.; Carmeli, S. Eight novel serine proteases inhibitors from a water bloom of the cyanobacterium Microcystis sp. Tetrahedron 2010, 66, 9194–9202.
- Matthew, S.; Ross, C.; Paul, V.J.; Luesch, H. Pompanopeptins A and B, new cyclic peptides from the marine cyanobacterium Lyngbya confervoides. Tetrahedron 2008, 64, 4081–4089.
- Sanz, M.; Andreote, A.P.D.; Fiore, M.F.; Dörr, F.A.; Pinto, E. Structural characterization of new peptide variants produced by cyanobacteria from the Brazilian Atlantic Coastal Forest using liquid chromatography coupled to quadrupole time-of-flight tandem mass spectrometry. Mar. Drugs 2015, 13, 3892–3919.
- Sano, T.; Usui, T.; Ueda, K.; Osada, H.; Kaya, K. Isolation of new protein phosphatase inhibitors from two cyanobacteria species, Planktothrix spp. J. Nat. Prod. 2001, 64, 1052–1055.
- Itou, Y.; Suzuki, S.; Ishida, K.; Murakami, M. Anabaenopeptins G and H, potent carboxypeptidase A inhibitors from the cyanobacterium Oscillatoria agardhii (NIES-595). Bioorganic Med. Chem. Lett. 1999, 9, 1243–1246.
- Tonk, L.; Welker, M.; Huisman, J.; Visser, P.M. Production of cyanopeptolins, anabaenopeptins, and microcystins by the harmful cyanobacteria Anabaena 90 and Microcystis PCC 7806. Harmful Algae 2009, 8, 219–224.
- Mi, Y.; Zhang, J.; He, S.; Yan, X. New Peptides Isolated from Marine Cyanobacteria, an Overview over the Past Decade. Mar. Drugs 2017, 15, 132.
- Fujii, K.; Harada, K.; Suzuki, M.; Kondo, F.; Ikai, Y.; Oka, H.; Sivonen, K. Novel cyclic peptides together with microcystins produced by toxic cyanobacteria, Anabaena sp. Symp. Chem. Nat. Prod. Symp. Pap. 1995, 37, 445–450.
- Greunke, C.; Duell, E.R.; D’Agostino, P.M.; Glöckle, A.; Lamm, K.; Gulder, T.A.M. Direct Pathway Cloning (DiPaC) to unlock natural product biosynthetic potential. Metab. Eng. 2018, 47, 334–345.
- Shin, H.J.; Matsuda, H.; Murakami, M.; Yamaguchi, K. Anabaenopeptins E and F, two new cyclic peptides from the cyanobacterium Oscillatoria agardhii (NIES-204). J. Nat. Prod. 1997, 60, 139–141.
- Murakami, M.; Suzuki, S.; Itou, Y.; Kodani, S.; Ishida, K. New Anabaenopeptins, Potent Carboxypeptidase-A Inhibitors from the Cyanobacterium Aphanizomenon flos-aquae. J. Nat. Prod. 2000, 63, 1280–1282.
- Kodani, S.; Suzuki, S.; Ishida, K.; Murakami, M. Five new cyanobacterial peptides from water bloom materials of lake Teganuma (Japan). FEMS Microbiol. Lett. 1999, 178, 343–348.
- Campos, A.; Vasconcelos, V. Molecular mechanisms of microcystin toxicity in animal cells. Int. J. Mol. Sci. 2010, 11, 268–287.
- Spoof, L.; Błaszczyk, A.; Meriluoto, J.; Cegłowska, M.; Mazur-Marzec, H. Structures and Activity of New Anabaenopeptins Produced by Baltic Sea Cyanobacteria. Mar. Drugs 2015, 14, 8.
- Erhard, M.; Von Döhren, H.; Jungblut, P.R. Rapid identification of the new anabaenopeptin G from Planktothrix agardhii HUB 011 using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1999, 13, 337–343.
- Harms, H.; Kurita, K.L.; Pan, L.; Wahome, P.G.; He, H.; Kinghorn, A.D.; Carter, G.T.; Linington, R.G. Discovery of anabaenopeptin 679 from freshwater algal bloom material: Insights into the structure–activity relationship of anabaenopeptin protease inhibitors. Bioorganic Med. Chem. Lett. 2016, 26, 4960–4965.
- Grabowska, M.; Kobos, J.; Toruńska-Sitarz, A.; Mazur-Marzec, H. Non-ribosomal peptides produced by Planktothrix agardhii from Siemianówka Dam Reservoir SDR (northeast Poland). Arch. Microbiol. 2014, 196, 697–707.
- de Carvalho, L.R.; Pipole, F.; Werner, V.R.; Laughinghous, H.D., IV; de Camargo, A.C.M.; Rangel, M.; Konno, K.; Sant’ Anna, C.L. A toxic cyanobacterial bloom in an urban coastal lake, Rio Grande do Sul state, Southern Brazil. Braz. J. Microbiol. 2008, 39, 761–769.
- Teta, R.; Della Sala, G.; Glukhov, E.; Gerwick, L.; Gerwick, W.H.; Mangoni, A.; Costantino, V. Combined LC–MS/MS and Molecular Networking Approach Reveals New Cyanotoxins from the 2014 Cyanobacterial Bloom in Green Lake, Seattle. Environ. Sci. Technol. 2015, 49, 14301–14310.
- Beversdorf, L.; Weirich, C.; Bartlett, S.; Miller, T. Variable Cyanobacterial Toxin and Metabolite Profiles across Six Eutrophic Lakes of Differing Physiochemical Characteristics. Toxins 2017, 9, 62.
- Bartlett, S.L.; Brunner, S.L.; Klump, J.V.; Houghton, E.M.; Miller, T.R. Spatial analysis of toxic or otherwise bioactive cyanobacterial peptides in Green Bay, Lake Michigan. J. Great Lakes Res. 2018, 44, 924–933.
- Roy-Lachapelle, A.; Solliec, M.; Sauvé, S.; Gagnon, C. A Data-Independent Methodology for the Structural Characterization of Microcystins and Anabaenopeptins Leading to the Identification of Four New Congeners. Toxins 2019, 11, 619.
- Flores, C.; Caixach, J. High Levels of Anabaenopeptins Detected in a Cyanobacteria Bloom from N.E. Spanish Sau-Susqueda-El Pasteral Reservoirs System by LC–HRMS. Toxins 2020, 12, 541.
- Kust, A.; Řeháková, K.; Vrba, J.; Maicher, V.; Mareš, J.; Hrouzek, P.; Chiriac, M.-C.; Benedová, Z.; Tesařová, B.; Saurav, K. Insight into Unprecedented Diversity of Cyanopeptides in Eutrophic Ponds Using an MS/MS Networking Approach. Toxins 2020, 12, 561.
- Riba, M.; Kiss-Szikszai, A.; Gonda, S.; Boros, G.; Vitál, Z.; Borsodi, A.K.; Krett, G.; Borics, G.; Ujvárosi, A.Z.; Vasas, G. Microcystis Chemotype Diversity in the Alimentary Tract of Bigheaded Carp. Toxins 2019, 11, 288.
- Konkel, R.; Toruńska-Sitarz, A.; Cegłowska, M.; Ežerinskis, Ž.; Šapolaitė, J.; Mažeika, J.; Mazur-Marzec, H. Blooms of Toxic Cyanobacterium Nodularia spumigena in Norwegian Fjords During Holocene Warm Periods. Toxins 2020, 12, 257.
- Logares, R.; Bråte, J.; Bertilsson, S.; Clasen, J.L.; Shalchian-Tabrizi, K.; Rengefors, K. Infrequent marine-freshwater transitions in the microbial world. Trends Microbiol. 2009, 17, 414–422.
- Zi, J.; Lantvit, D.D.; Swanson, S.M.; Orjala, J. Lyngbyaureidamides A and B, two anabaenopeptins from the cultured freshwater cyanobacterium Lyngbya sp. (SAG 36.91). Phytochemistry 2012, 74, 173–177.
- Williams, D.E.; Craig, M.; Holmes, C.F.B.; Andersen, R.J. Ferintoic acids A and B, new cyclic hexapeptides from the freshwater cyanobacterium Microcystis aeruginosa. J. Nat. Prod. 1996, 59, 570–575.
- Natumi, R.; Janssen, E.M.L. Cyanopeptide Co-Production Dynamics beyond Mirocystins and Effects of Growth Stages and Nutrient Availability. Environ. Sci. Technol. 2020, 54, 6063–6072.
- Gesner-Apter, S.; Carmeli, S. Protease Inhibitors from a Water Bloom of the Cyanobacterium Microcystis aeruginosa. J. Nat. Prod. 2009, 72, 1429–1436.
- Beresovsky, D.; Hadas, O.; Livne, A.; Sukenik, A.; Kaplan, A.; Carmeli, S. Toxins and Biologically Active Secondary Metabolites of Microcystis sp. isolated from Lake Kinneret. Isr. J. Chem. 2006, 46, 79–87.
- Elkobi-Peer, S.; Carmeli, S. New Prenylated Aeruginosin, Microphycin, Anabaenopeptin and Micropeptin Analogues from a Microcystis Bloom Material Collected in Kibbutz Kfar Blum, Israel. Mar. Drugs 2015, 13, 2347–2375.
- Mazur-Marzec, H.; Bertos-Fortis, M.; Toruńska-Sitarz, A.; Fidor, A.; Legrand, C. Chemical and genetic diversity of nodularia spumigena from the baltic sea. Mar. Drugs 2016, 14, 209.
- Okumura, H.S.; Philmus, B.; Portmann, C.; Hemscheidt, T.K. Homotyrosine-Containing Cyanopeptolins 880 and 960 and Anabaenopeptins 908 and 915 from Planktothrix agardhii CYA 126/8. J. Nat. Prod. 2009, 72, 172–176.
- Tooming-Klunderud, A.; Sogge, H.; Rounge, T.B.; Nederbragt, A.J.; Lagesen, K.; Glöckner, G.; Hayes, P.K.; Rohrlack, T.; Jakobsen, K.S. From Green to Red: Horizontal Gene Transfer of the Phycoerythrin Gene Cluster between Planktothrix Strains. Appl. Environ. Microbiol. 2013, 79, 6803–6812.
- Welker, M.; Erhard, M. Consistency between chemotyping of single filaments ofPlanktothrix rubescens (cyanobacteria) by MALDI-TOF and the peptide patterns of strains determined by HPLC-MS. J. Mass Spectrom. 2007, 42, 1062–1068.
- Rohrlack, T.; Edvardsen, B.; Skulberg, R.; Halstvedt, C.B.; Utkilen, H.C.; Ptacnik, R.; Skulberg, O.M. Oligopeptide chemotypes of the toxic freshwater cyanobacterium Planktothrix can form sub-populations with dissimilar ecological traits. Limnol. Oceanogr. 2008, 53, 1279–1293.
- Sedmak, B.; Carmeli, S.; Eleršek, T. “Non-Toxic” Cyclic Peptides Induce Lysis of Cyanobacteria—An Effective Cell Population Density Control Mechanism in Cyanobacterial Blooms. Microb. Ecol. 2008, 56, 201–209.
- Grach-Pogrebinsky, O.; Sedmak, B.; Carmeli, S. Protease inhibitors from a Slovenian Lake Bled toxic waterbloom of the cyanobacterium Planktothrix rubescens. Tetrahedron 2003, 59, 8329–8336.
- Vasas, G.; Farkas, O.; Borics, G.; Felföldi, T.; Sramkó, G.; Batta, G.; Bácsi, I.; Gonda, S. Appearance of Planktothrix rubescens Bloom with [D-Asp3, Mdha7]MC–RR in Gravel Pit Pond of a Shallow Lake-Dominated Area. Toxins 2013, 5, 2434–2455.
- Bober, B.; Chrapusta-Srebrny, E.; Bialczyk, J. Novel cyanobacterial metabolites, cyanopeptolin 1081 and anabaenopeptin 899, isolated from an enrichment culture dominated by Woronichinia naegeliana (Unger) Elenkin. Eur. J. Phycol. 2020, 1–11.
- Grach-Pogrebinsky, O.; Carmeli, S. Three novel anabaenopeptins from the cyanobacterium Anabaena sp. Tetrahedron 2008, 64, 10233–10238.
- Häggqvist, K.; Toruńska-Sitarz, A.; Błaszczyk, A.; Mazur-Marzec, H.; Meriluoto, J. Morphologic, Phylogenetic and Chemical Characterization of a Brackish Colonial Picocyanobacterium (Coelosphaeriaceae) with Bioactive Properties. Toxins 2016, 8, 108.
- Fujii, K.; Sivonen, K.; Adachi, K.; Noguchi, K.; Sano, H.; Hirayama, K.; Suzuki, M.; Harada, K. Comparative study of toxic and non-toxic cyanobacterial products: Novel peptides from toxic Nodularia spumigena AV1. Tetrahedron Lett. 1997, 38, 5525–5528.
- Popin, R.V.; Delbaje, E.; de Abreu, V.A.C.; Rigonato, J.; Dörr, F.A.; Pinto, E.; Sivonen, K.; Fiore, M.F. Genomic and Metabolomic Analyses of Natural Products in Nodularia spumigena Isolated from a Shrimp Culture Pond. Toxins 2020, 12, 141.
- Saha, S.; Esposito, G.; Urajová, P.; Mareš, J.; Ewe, D.; Caso, A.; Macho, M.; Delawská, K.; Kust, A.; Hrouzek, P.; et al. Discovery of Unusual Cyanobacterial Tryptophan-Containing Anabaenopeptins by MS/MS-Based Molecular Networking. Molecules 2020, 25, 3786.
- Nowruzi, B.; Khavari-Nejad, R.-A.; Sivonen, K.; Kazemi, B.; Najafi, F.; Nejadsattari, T. Identification and toxigenic potential of a Nostoc sp. Algae 2012, 27, 303–313.
- Reshef, V.; Carmeli, S. Schizopeptin 791, a new anabeanopeptin-like cyclic peptide from the cyanobacterium Schizothrix sp. J. Nat. Prod. 2002, 65, 1187–1189.
- Entfellner, E.; Frei, M.; Christiansen, G.; Deng, L.; Blom, J.; Kurmayer, R. Evolution of Anabaenopeptin Peptide Structural Variability in the Cyanobacterium Planktothrix. Front. Microbiol. 2017, 8.
- Mutalipassi, M.; Riccio, G.; Mazzella, V.; Galasso, C.; Somma, E.; Chiarore, A.; de Pascale, D.; Zupo, V. Symbioses of Cyanobacteria in Marine Environments: Ecological Insights and Biotechnological Perspectives. Mar. Drugs 2021, 19, 227.
- Carpenter, E.J.; Foster, R.A. Marine cyanobacteria symbioses. In Cyanobacteria in Symbiosis; Rai, A.N., Bergman, B., Rasmussen, U., Eds.; Kluwer Academic Publisher: Dordrecht, The Netherlands, 2002; pp. 10–17. ISBN 0306480050.
- Shridhar, D.M.P.; Mahajan, G.; Kamat, V.; Naik, C.; Parab, R.; Thakur, N.; Mishra, P. Antibacterial Activity of 2-(2′,4′-Dibromophenoxy)-4,6-dibromophenol from Dysidea granulosa. Mar. Drugs 2009, 7, 464–471.
- Plaza, A.; Keffer, J.L.; Lloyd, J.R.; Colin, P.L.; Bewley, C.A. Paltolides A−C, Anabaenopeptin-Type Peptides from the Palau Sponge Theonella swinhoei. J. Nat. Prod. 2010, 73, 485–488.
- Kobayashi, J.; Sato, M.; Murayama, T.; Ishibashi, M.; Wälchi, M.R.; Kanai, M.; Shoji, J.; Ohizumi, Y. Konbamide, a novel peptide with calmodulin antagonistic activity from the Okinawan marine sponge Theonella sp. J. Chem. Soc. Chem. Commun. 1991, 1050–1052.
- Robinson, S.J.; Tenney, K.; Yee, D.F.; Martinez, L.; Media, J.E.; Valeriote, F.A.; van Soest, R.W.M.; Crews, P. Probing the Bioactive Constituents from Chemotypes of the Sponge Psammocinia aff. bulbosa. J. Nat. Prod. 2007, 70, 1002–1009.
- Mazur-Marzec, H.; Błaszczyk, A.; Felczykowska, A.; Hohlfeld, N.; Kobos, J.; Toruńska-Sitarz, A.; Devi, P.; Montalvão, S.; D’souza, L.; Tammela, P.; et al. Baltic cyanobacteria—A source of biologically active compounds. Eur. J. Phycol. 2015, 50, 343–360.
- Gulledge, B.; Aggen, J.; Huang, H.; Nairn, A.; Chamberlin, A. The Microcystins and Nodularins: Cyclic Polypeptide Inhibitors of PP1 and PP2A. Curr. Med. Chem. 2002, 9, 1991–2003.
- Shah, S.; Akhter, N.; Auckloo, B.; Khan, I.; Lu, Y.; Wang, K.; Wu, B.; Guo, Y.-W. Structural Diversity, Biological Properties and Applications of Natural Products from Cyanobacteria. A Review. Mar. Drugs 2017, 15, 354.
- Wahome, P.; Beauchesne, K.; Pedone, A.; Cavanagh, J.; Melander, C.; Zimba, P.; Moeller, P. Augmenting Anti-Cancer Natural Products with a Small Molecule Adjuvant. Mar. Drugs 2014, 13, 65–75.
- Shishido, T.K.; Popin, R.V.; Jokela, J.; Wahlsten, M.; Fiore, M.F.; Fewer, D.P.; Herfindal, L.; Sivonen, K. Dereplication of Natural Products with Antimicrobial and Anticancer Activity from Brazilian Cyanobacteria. Toxins 2019, 12, 12.