
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Lili Wan | + 2262 word(s) | 2262 | 2021-07-22 16:26:31 | | | |
| 2 | Conner Chen | Meta information modification | 2262 | 2021-08-04 14:13:00 | | |
Video Upload Options
Fruit and vegetable crops are rich in dietary fibre, vitamins and minerals, which are vital to human health. However, many biotic stressors (such as pests and diseases) and abiotic stressors threaten crop growth, quality, and yield. Traditional breeding strategies for improving crop traits include a series of backcrosses and selection to introduce beneficial traits into fine germplasm, this process is slow and resource-intensive. The new breeding technique known as clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein-9 (Cas9) has the potential to improve many traits rapidly and accurately, such as yield, quality, disease resistance, abiotic stress tolerance, and nutritional aspects in crops. Because of its simple operation and high mutation efficiency, this system has been applied to obtain new germplasm resources via gene-directed mutation.
1. The Discovery and Development of CRISPR Technology
CRISPR-associated (Cas) genes were first discovered in the Escherichia coli genome in 1987 and were officially named by the Dutch scientist who identified them [1]. In 2005, it was discovered that many CRISPR spacers consist of short sequences that are highly homologous with sequences originating from extrachromosomal DNA. The Cas-encoded protein can combine with the CRISPR transcription products and with the homologous foreign DNA sequences to form a protein–RNA complex, which can cut the foreign DNA fragments. The primary function of the CRISPR complex in bacteria and archaea is to integrate specific fragments of exogenous DNA (from invading phages or other sources) into their own genomes to become interval sequences. During subsequent invasion by foreign DNA, the specific recognition system is then activated, providing an acquired immune defence function [2][3][4].
CRISPR-Cas technology has been successfully applied to the editing of human, animal, and plant genomes, and has been developed for use in drug screening, animal domestication, and food science research [5][6][7]. There are three main types of CRISPR-Cas systems. Types I and III use a large multi-Cas protein complex for interference [8]. Type II requires only a simple effector-module architecture to accomplish interference via its two signature nuclease domains, RuvC and HNH [9]. Among various CRISPR nucleases, type II Cas9 from Streptococcus pyogenes (SpCas9) is the most widely used in CRISPR-Cas technology [10]. The sgRNA-Cas complex recognises the protospacer adjacent motif (PAM) and Cas9 cleaves the target DNA to generate a double-strand break (DSB), triggering cellular DNA repair mechanisms (Figure 1). In eukaryotes, DSBs have two main repair mechanisms. The first is nonhomologous end joining (NHEJ). In the absence of a homologous repair template, the NHEJ repair pathway is activated at the DSB site, thus disrupting gene function. The second is homology directed repair (HDR). If a donor DNA template homologous to the sequence surrounding the DSB site is available, the HDR pathway is initiated, precisely introducing specific mutations such as insertion or replacement of desired sequences into the break sites [11]. Using a donor DNA as a template, gene targeting (GT) can precisely modify a target locus to repair DNA DSBs.
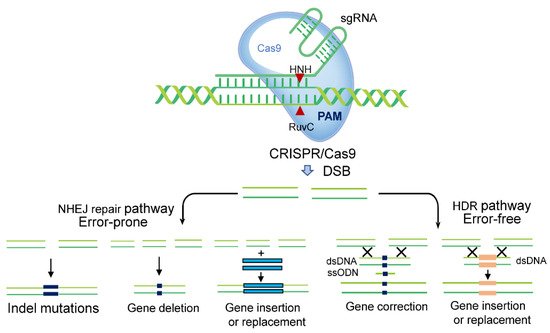
Figure 1. The potential applications of CRISPR-Cas systems in genome editing. CRISPR-Cas systems mediated genome modification depending on the two main double-strand break (DSB) repair pathways. Indel mutation and gene deletion are outcomes of the dominant nonhomologous end joining (NHEJ) repair pathway. Gene insertion, correction, and replacement, using a DNA donor template, are outcomes of the homology directed repair (HDR) pathway.
Several strategies are used to improve the homologous recombination frequency between a genomic target and an exogenous homologous template donor. Most of the strategies focus on enhancing the number of donor repair templates using virus replicons [12], suppressing the NHEJ pathway [13], and timing DSB induction at target sites to coincide with donor repair template delivery in plant cells [14]. Finally, the recombination frequency can be enhanced by treatment with Rad51-stimulatory compound1 (RS-1) [15].
2. CRISPR-Cas9 in Fruit and Vegetable Crop Improvement
In 2014, CRSPR-Cas9 was used to create the first needle-leaf mutant in tomato, by knocking out Argonaute 7 [16]. Many studies have since been published on its possible applications in protecting plants against biotic and abiotic stresses, and improving fruit quality, plant architecture, and shelf life [17]. Currently, the system is in the research stage for many fruits and vegetables crops, such as cabbage, mustard, tomato, and watermelon.
Most gene-editing studies have evaluated mutation efficiency in terms of the number of albino plants obtained after mutation of the endogenous phytoene desaturase (PDS) gene. The disruption of PDS impairs the production of chlorophyll and carotenoid, generating an easily identifiable albinism phenotype in plants. However, the products of gene editing obtained in this way have no economic value [18][19][20]. Because of its high economic value and the availability of Agrobacterium-mediated transformation, tomato has become a model crop for testing CRISPR-Cas9 applications (Figure 2).
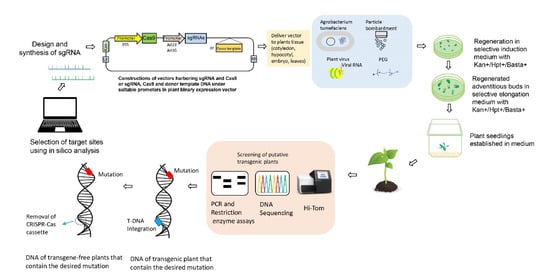
Figure 2. CRISPR-Cas9 mediated genome editing. (I) Selection of the desired genomic DNA target, and recognition of protospacer adjacent motif (PAM) sequences before 20 bp sequences. Design of the sgRNA using online bioinformatics tools. (II) Cloning of designed sgRNAs, and binary vector construction using promoters. (III) The delivery of CRISPR-Cas editing reagents into plant cells. The vector can be transferred into the plant via Agrobacterium tumefaciens, nanoparticles, biolistic bombardment, or polyethylene glycol (PEG). Alternatively, plant RNA viruses have been used to induce heritable genome editing. When the cassette harbouring the sgRNA, RNA mobile element, and tobacco rattle virus (TRV) is transformed into the Cas9 expressing plants, the systemic spread of sgRNA will introduce heritable genome editing. (IV) Plant transformation and development of transgenic plants. (V) Genotyping of transgenic plants. (VI) Transgene-free plants with the desired mutation are obtained.
2.1. Improvement of Biotic Stress Resistance
Two strategies have been used to improve plant resistance to viruses: (1) designing sgRNAs and targeting the virus genome; or (2), modifying the fruit crop genes in the antiviral pathway. The binding of virus genome linked protein (VPg) to the plant protein ‘eukaryotic translation initiation factor 4E’ (eIF4E) is key in Y virus infection of plants. Mutation of a key site of eIF4E can affect the virus–plant interaction, and mediate plant resistance to this virus [21]. In cucumbers, using CRISPR-Cas to target the N′ and C′ ends of eIF4E-produced nontransgenic homozygous plants in the T3 generation; these showed immunity to cucumber vein yellow virus and pumpkin mosaic virus, and resistance to papaya ring spot mosaic virus (PRSV-W) [22].
CRISPR-Cas9 can generate mutations in the coding and noncoding regions of geminivirus, effectively reducing its pathogenicity. In Nicotiana benthamiana, sgRNA-Cas9 constructs target beet severe curly top virus (a geminivirus), inhibiting its accumulation in leaves [23]. Geminivirus noncoding-region mutations are believed to reduce or even inhibit its replication ability. Compared with coding-region mutations, noncoding-region mutations generate fewer viral variants [24].
Fungi cause many diseases, potentially causing severe losses in crop yield and quality. For instance, downy and powdery mildews cause serious economic losses in tomato [25]. Arabidopsis thaliana DMR6 (down mildew resistant) is a member of the 2-oxoglutarate oxygenase Fe(II)-dependent superfamily and is involved in salicylic acid homeostasis. Overexpression of DMR6 in plants can reduce susceptibility to downy mildew [26]. The DMR6 mutation obtained using CRISPR-Cas9 to knock out the homologous genes in tomato showed resistance to Pseudomonas syringae, Phytophthora and Xanthomonas spp. [27]. Mlo1 (Mildew resistant locus 1) encodes a membrane-associated protein and is a powdery mildew disease-sensitivity gene. In tomato, Mlo1 mutants obtained via gene editing exhibited resistance to the powdery mildew Oidium neolycopersici. Further, a mutant free of mlo1 T-DNA was obtained by selfing T0 generation plants [28].
The fungal pathogen Fusarium oxysporum can cause Fusarium wilt disease in fruit and vegetable crops [29]. In tomatoes, Solyc08g075770-knockout via CRISPR-Cas9 resulted in sensitivity to Fusarium wilt disease [30]. In watermelons, the knockout of Clpsk1, encoding the Phytosulfokine (PSK) precursor, confers enhanced resistance to Fusarium oxysporum f.sp.niveum (FON) [31]. Botrytis cinerea, an airborne plant pathogen that infects fruit and vegetable crops, causes great economic losses. Its initial symptoms are not obvious, and the lack of effective pesticides makes its prevention and control difficult. Pathogens can be effectively controlled in crops by the use of genetic resources that convey heritable resistance. In tomatoes, mutations in MAPK3 (mitogen-activated protein kinase 3) produced using CRISPR-Cas9 induce resistance to Botrytis cinerea [32].
The bacterial pathogen Pseudomonas syringae causes leaf spot diseases in crops, severely impacting the yield and sensory qualities of fruits and vegetables. In Arabidopsis thaliana, CRISPR-Cas9 was used to mutate the C-terminal jasmonate domain (JAZ2Δjas) of JAZ2 (jasmonate ZIM domain protein 2), causing expression of JAZ2 repressors; these repressors confer resistance to Pseudomonas syringae [33].
2.2. Abiotic Stress Resistance Improvement
With climate change, crop production is exposed to increased potential risks of abiotic stress. Although traditional breeding can to some extent ensure stable crop production, the application of new technologies to rapidly obtain new crop germplasm resources capable of responding to abiotic stress is essential for accelerating the cultivation of new varieties [34]. The emergence of CRISPR-Cas9 gene editing has shortened the time required to create new varieties. Brassinazole-resistant 1 gene (BZR1) participates in various brassinosteroid (BR) mediated development processes. The CRISPR mediated mutation in BZR1 impaired the induction of RESPIRATORY BURST OXIDASE HOMOLOG1(RBOH1) and the production of H2O2. Exogenous H2O2 recovered the heat tolerance in tomato bzr1 mutant [35]. Further, new cold- and drought-tolerant germplasms can be created using gene-editing, for instance, of CBF1 (C-repeat binding factor 1), which regulates cold tolerance in plants, and MAPK3, which participates in the drought stress response to protect plant cell membranes from peroxidative damage in tomatoes [36][37].
2.3. Herbicide Resistance Improvement
Weeds are an important cause of stress that affect vegetable yield and quality, and selective herbicides are often used to control weed growth during cultivation. To obtain herbicide-resistant fruits and vegetables for field production, CRISPR-Cas9 gene editing was used for site-directed mutagenesis of the herbicide target gene acetolactate synthase (ALS) in watermelon, yielding a herbicide-resistant watermelon germplasm [38]. Cytidine base editing (CBE) was used for cytidine editing of key ALS sites in tomato and potato, resulting in amino acid mutations. Up to 71% of edited tomato plants exhibited resistance to the pesticide chlorsulfuron, and of the edited tomato and potato plants, 12% and 10%, respectively, were free of GM components [39]. Phelipanche aegyptiaca, an obligate weedy plant parasite, requires the host roots to release the plant hormone strigolactone (SL) to promote seed germination; CRISPR-Cas9 was used to mutate carotenoid dioxygenase 8 (CCD8), a key enzyme in the carotenoid synthesis pathway that produces SLs in tomato, and More Axillary Growth1 (MAX1), which is involved in the synthesis of SLs, thereby significantly reducing SL content, and creating P. aegyptiaca-resistant tomato plants [40][41].
2.4. Fruit and Vegetable Quality Improvement
The primary goal in fruit and vegetable breeding is to improve quality and prolong shelf life after harvest. Quality refers to both external and internal factors. External quality refers to fruit size, colour, and texture, which can be discerned by the naked eye. Internal quality must be measured using equipment, and includes the levels of nutrients such as sugars, vitamins, and bioactive compounds including lycopene, anthocyanins, and malate. For example, in tomato, the ovary locule number, which determines 50% of the genetic variation in fruit size, is determined by multiple QTLs [42]. Researchers at Cold Spring Harbor Laboratory designed eight sgRNAs and used CRISPR-Cas9 to edit the promoter region of the tomato CLAVATA-WUSCHEL (CLV-WUS) stem cell gene CLV3 to obtain fruits that are larger and more numerous than wild-type fruits [43]; editing of fruit-size determining QTLs, such as the QTLs for locule number (lc) and fasciated number (fas), generated germplasm resources with an increased number of locules [44].
Fruit and vegetable colour and texture are important traits for consumers. For example, European and American consumers prefer red tomatoes, whereas Asian consumers prefer pink tomatoes [45][46]. CRISPR-Cas was used to modify phytoene synthase 1 (PSY1), MYB transcription factor 12 (MYB12), and anthocyanin 2 (ANT2) to obtain yellow, pink, and purple tomatoes, respectively [47][48][49]. The carotenoid isomerase gene of Chinese kale (BoaCRTISO) is responsible for catalysis, then conversion of lycopene precursors to lycopene. When BoaCRTISO was targeted and edited, the colour of mutants changed from green to yellow [50]. The primary goal of improving the intrinsic quality of fruits and vegetables is to improve their nutrient and bioactive compound content. Carbohydrates and vitamins are essential nutrients. Many genes are involved in the synthesis and metabolism of sucrose and carotenoids. One of the carotenoids, provitamin A, can be absorbed by the human body and converted into vitamin A. For example, CRISPR-Cas was used to knock out MPK20 (mitogen-activated protein kinase 20), blocking the transcription and protein products of multiple genes in the sucrose metabolism pathway [51]. Biofortification, the biotechnological improvement of the absorption, transport, and metabolism of minerals by plants, increases the levels of micronutrients that are beneficial to human health; long-term consumption of these micronutrients can effectively prevent cardiovascular disease and cancer [52].
Anthocyanins [53], malate [54], γ-aminobutyric acid (GABA) [55], and lycopene [56] are bioactive compounds. Adjusting key metabolic-pathway-related genes via CRISPR-Cas9 can enrich these nutrients in fruits. For example, in tomatoes, butylamine content was increased 19-fold through editing multiple genes in the GABA synthesis pathway, and malate content was improved by regulating aluminium-activated malate transporter (ALMT9) [55].
CRISPR-Cas9 can also be used to reduce the content of substances in vegetables that are not conducive to human health, by targeting mutations that inactivate genes in biosynthetic pathways. In potato tubers, for example, excessive content of steroidal glycoalkaloids (SGAs), such as α-solanine and α-chaconine, affects their taste and makes them less safe for human consumption, hence low content is an indicator of high quality. CRISPR-Cas9 was used to delete St16DOX (steroid 16α-hydroxylase) in the potato SGA biosynthetic pathway, resulting in SGA-free potato lines [57].
Prolonged shelf-life is an important breeding goal in fruit and vegetable production. CRISPR was used to knock out ripening inhibitor (RIN) or DNA demethylase (DNA demethylase 2, DML2) to slow fruit ripening, thereby prolonging their shelf life. However, regulating these two genes in fruit alters peel colour and reduces flavour and nutritional value, severely reducing the fruit’s palatability and sensory qualities [58][59]. In tomatoes, inhibiting the expression of the pectate lyase (PL) and alcobaca (ALC) genes effectively extended shelf life, without affecting the sensory qualities or nutritional value [60][61].
References
- Jansen, R.; Embden, J.D.V.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575.
- Bolotin, A. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005, 151, 2551–2561.
- Mojica, F.J.; Díez-Villaseñor, C.; García-Martínez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182.
- Pourcel, C. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 2005, 151, 653–663.
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823.
- Kaboli, S.; Babazada, H. CRISPR Mediated Genome Engineering and its Application in Industry. Curr. Issues Mol. Biol. 2018, 26, 81–92.
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688.
- Rouillon, C.; Zhou, M.; Zhang, J.; Politis, A.; Beilsten-Edmands, V.; Cannone, G.; Graham, S.; Robinson, C.V.; Spagnolo, L.; White, M.F. Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol. Cell 2013, 52, 124–134.
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 15539–15540.
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096.
- Yin, K.; Gao, C.; Qiu, J.L. Progress and prospects in plant genome editing. Nat. Plants 2017, 3, 17107.
- Baltes, N.J.; Gil-Humanes, J.; Cermak, T.; Atkins, P.A.; Voytas, D.F. DNA replicons for plant genome engineering. Plant cell 2014, 26, 151–163.
- Endo, M.; Ishikawa, Y.; Osakabe, K.; Nakayama, S.; Kaya, H.; Araki, T.; Shibahara, K.-I.; Abe, K.; Ichikawa, H.; Valentine, L.; et al. Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 2006, 25, 5579–5590.
- Gil-Humanes, J.; Wang, Y.; Liang, Z.; Shan, Q.; Ozuna, C.V.; Sánchez-León, S.; Baltes, N.J.; Starker, C.; Barro, F.; Gao, C.; et al. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 2017, 89, 1251–1262.
- Ayathilaka, K.; Sheridan, S.D.; Bold, T.D.; Bochenska, K.; Logan, H.L.; Weichselbaum, R.R.; Bishop, D.K.; Connell, P.P. A chemical compound that stimulates the human homologous recombination protein RAD51. Proc. Natl. Acad. Sci. USA 2008, 105, 15848–15853.
- Brooks, C.; Nekrasov, V.; Lippman, Z.B.; Van Eck, J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014, 166, 1292–1297.
- Kulus, D. Genetic resources and selected conservation methods of tomato. J. Appl. Bot. Food Qual. 2018, 91, 135–144.
- Ma, C.; Liu, M.; Li, Q.; Si, J.; Ren, X.; Song, H. Efficient BoPDS Gene Editing in Cabbage by the CRISPR/Cas9 System. Hortic. Plant J. 2019, 5, 164–169.
- Sun, B.; Zheng, A.; Jiang, M.; Xue, S.; Yuan, Q.; Jiang, L.; Chen, Q.; Li, M.; Wang, Y.; Zhang, Y.; et al. CRISPR/Cas9-mediated mutagenesis of homologous genes in Chinese kale. Sci. Rep. 2018, 8, 16786.
- Tian, S.; Jiang, L.; Gao, Q.; Zhang, J.; Zong, M.; Zhang, H.; Ren, Y.; Guo, S.; Gong, G.; Liu, F.; et al. Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 2017, 36, 399–406.
- Bastet, A.; Zafirov, D.; Giovinazzo, N.; Guyon-Debast, A.; Nogué, F.; Robaglia, C.; Gallois, J.L. Mimicking natural polymorphism in eIF4E by CRISPR-Cas9 base editing is associated with resistance to potyviruses. Plant Biotechnol. J. 2019, 17, 1736–1750.
- Chandrasekaran, J.; Brumin, M.; Wolf, D.; Leibman, D.; Klap, C.; Pearlsman, M.; Sherman, A.; Arazi, T.; Gal-On, A. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 2016, 17, 1140–1153.
- Ji, X.; Zhang, H.; Zhang, Y.; Wang, Y.; Gao, C. Establishing a CRISPR–Cas-like immune system conferring DNA virus resistance in plants. Nat. Plants 2015, 1, 15144.
- Ali, Z.; Ali, S.; Tashkandi, M.; Zaidi, S.S.E.A.; Mahfouz, M.M. CRISPR/Cas9-Mediated Immunity to Geminiviruses: Differential Interference and Evasion. Sci. Rep. 2016, 6, 26912.
- Borrelli, V.M.G.; Brambilla, V.; Rogowsky, P.; Marocco, A.; Lanubile, A. The Enhancement of Plant Disease Resistance Using CRISPR/Cas9 Technology. Frontiers in Plant Science. 2018, 9, 1245.
- Zeilmaker, T.; Ludwig, N.R.; Elberse, J.; Seidl, M.F.; Berke, L.; Van Doorn, A.; Schuurink, R.C.; Snel, B.; Van den Ackerveken, G. DOWNY MILDEW RESISTANT 6 and DMR6-LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J. 2015, 81, 210–222.
- Paula de Toledo Thomazella, D.; Brail, Q.; Dahlbeck, D.; Staskawicz, B. CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. BioRxiv 2016, 064824.
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 2017, 7, 1–6.
- Chaudhary, R.; Hs, A. Resistance-Gene-Mediated Defense Responses against Biotic Stresses in the Crop Model Plant Tomato. J. Plant Pathol. Microbiol. 2017, 8, 2.
- Cahya, P.; Barbetti, M.J.; Barker, S.J. A Novel Tomato Fusarium Wilt Tolerance Gene. Front. Microbiol. 2018, 9, 1226.
- Zhang, M.; Liu, Q.; Yang, X.; Xu, J.; Liu, G.; Yao, X.; Ren, R.; Xu, J.; Lou, L. CRISPR/Cas9-mediated mutagenesis of Clpsk1 in watermelon to confer resistance to Fusarium oxysporum f.sp. niveum. Plant Cell Rep. 2020, 39, 589–595.
- Shujuan, Z.; Liu, W.; Ruirui, Z.; Wenqing, Y.; Rui, L.; Yujing, L.; Jiping, S.; Lin, S. Knockout of SlMAPK3 Reduced Disease Resistance to Botrytis cinerea in Tomato Plants. J. Agric. Food Chem. 2018, 66, 8949–8956.
- Andrés, O.; Selena, G.I.; Nathalie, L.; Roberto, S. Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol. J. 2018, 17, 665–673.
- Haque, E.; Taniguchi, H.; Hassan, M.M.; Bhowmik, P.; Karim, M.R.; Śmiech, M.; Zhao, K.; Rahman, M.; Islam, T. Application of CRISPR/Cas9 Genome Editing Technology for the Improvement of Crops Cultivated in Tropical Climates: Recent Progress, Prospects, and Challenges. Front. Plant Sci. 2018, 9, 617.
- Yin, Y.; Qin, K.; Song, X.; Zhang, Q.; Zhou, Y.; Xia, X.; Yu, J. BZR1 Transcription Factor Regulates Heat Stress Tolerance Through FERONIA Receptor-Like Kinase-Mediated Reactive Oxygen Species Signaling in Tomato. Plant Cell Physiol. 2018, 59, 2239–2254.
- Li, R.; Zhang, L.; Wang, L.; Chen, L.; Zhao, R.; Sheng, J.; Shen, L. Reduction of Tomato-Plant Chilling Tolerance by CRISPR-Cas9-Mediated SlCBF1 Mutagenesis. J. Agric. Food Chem. 2018, 66, 9042–9051.
- Wang, L.; Chen, L.; Li, R.; Zhao, R.; Yang, M.; Sheng, J.; Shen, L. Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J. Agric. Food Chem. 2017, 65, 8674–8682.
- Tian, S.; Jiang, L.; Cui, X.; Zhang, J.; Guo, S.; Li, M.; Zhang, H.; Ren, Y.; Gong, G.; Zong, M. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 2018, 37, 1353–1356.
- Butler, N.M.; Atkins, P.A.; Voytas, D.F.; Douches, D.S. Generation and Inheritance of Targeted Mutations in Potato (Solanum tuberosum L.) Using the CRISPR/Cas System. PLoS ONE 2015, 10, e0144591.
- Bari, V.K.; Nassar, J.A.; Aly, R. CRISPR/Cas9 mediated mutagenesis of MORE AXILLARY GROWTH 1 in tomato confers resistance to root parasitic weed Phelipanche aegyptiaca. Sci. Rep. 2021, 11, 3905.
- Bari, V.K.; Nassar, J.A.; Kheredin, S.M.; Gal-On, A.; Ron, M.; Britt, A.; Steele, D.; Yoder, J.; Aly, R. CRISPR/Cas9-mediated mutagenesis of CAROTENOID CLEAVAGE DIOXYGENASE 8 in tomato provides resistance against the parasitic weed Phelipanche aegyptiaca. Sci. Rep. 2019, 9, 11438.
- Li, H.; Qi, M.; Sun, M.; Liu, Y.; Liu, Y.; Xu, T.; Li, Y.; Li, T. Tomato Transcription Factor SlWUS Plays an Important Role in Tomato Flower and Locule Development. Front. Plant Sci. 2017, 8, 457.
- Li, T.; Yang, X.; Yu, Y.; Si, X.; Zhai, X.; Zhang, H.; Dong, W.; Gao, C.; Xu, C. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 2018, 36, 1160–1163.
- Rodríguez-Leal, D.; Lemmon, Z.H.; Man, J.; Bartlett, M.E.; Lippman, Z.B. Engineering Quantitative Trait Variation for Crop Improvement by Genome Editing. Cell 2017, 171, 470–480.
- Lin, T.; Zhu, G.; Zhang, J.; Xu, X.; Yu, Q.; Zheng, Z.; Zhang, Z.; Lun, Y.; Li, S.; Wang, X.; et al. Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 2014, 46, 1220–1226.
- Ballester, A.R.; Molthoff, J.; de Vos, R.; Hekkert, B.T.L.; Orzaez, D.; Fernaݩndez-Moreno, J.P.; Tripodi, P.; Gran-dillo, S.; Martin, C.; Heldens, J.; et al. Biochemical and Molecular Analysis of Pink Tomatoes: Deregulated Expression of the Gene Encoding Transcription Factors SlMYB12 Leads to Pink Tomato Fruit Color. Plant Physiol. 2010, 152, 71–84.
- Čermák, T.; Baltes, N.J.; Čegan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232.
- Deng, L.; Wang, H.; Sun, C.; Li, Q.; Jiang, H.; Du, M.; Li, C.B.; Li, C. Efficient generation of pink-fruited tomatoes using CRISPR/Cas9 system. J. Genet. Genom. 2018, 45, 51–54.
- Filler Hayut, S.; Melamed Bessudo, C.; Levy, A.A. Targeted recombination between homologous chromosomes for precise breeding in tomato. Nat. Commun. 2017, 8, 15605.
- Sun, B.; Jiang, M.; Zheng, H.; Jian, Y.; Huang, W.L.; Yuan, Q.; Zheng, A.H.; Chen, Q.; Zhang, Y.T.; Lin, Y.X.; et al. Color-related chlorophyll and carotenoid concentrations of Chinese kale can be altered through CRISPR/Cas9 targeted editing of the carotenoid isomerase gene BoaCRTISO. Hortic. Res. 2020, 7, 161.
- Chen, L.; Yang, D.; Zhang, Y. Evidence for a specific and critical role of mitogen-activated protein kinase 20 in uni-to-binucleate transition of microgametogenesis in tomato. Plant Biol. 2018, 219, 176–194.
- Kris-Etherton, P.M.; Hecker, K.D.; Bonanome, A.; Coval, S.M.; Binkoski, A.E.; Hilpert, K.F.; Griel, A.E.; Etherton, T.D. Bioactive compounds in foods: Their role in the prevention of cardiovascular disease and cancer. Am. J. Med. 2002, 113, 71–88.
- Meng, X.; Yang, D.; Li, X.; Zhao, S.; Sui, N.; Meng, Q. Physiological changes in fruit ripening caused by overexpression of tomato SlAN2, an R2R3-MYB factor. Plant Physiol. Biochem. 2015, 89, 24–30.
- Ye, J.; Wang, X.; Hu, T.; Zhang, F.; Wang, B.; Li, C.; Yang, T.; Li, H.; Lu, Y. An InDel in the Promoter of Al-ACTIVATED MALATE TRANSPORTER9 Selected during Tomato Domestication Determines Fruit Malate Contents and Aluminum Tolerance. Plant Cell 2017, 29, 2249.
- Nonaka, S.; Arai, C.; Takayama, M.; Matsukura, C.; Ezura, H. Efficient increase of γ-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci. Rep. 2017, 7, 7057.
- Li, X.; Wang, Y.; Chen, S.; Tian, H.; Fu, D.; Zhu, B.; Luo, Y.; Zhu, H. Lycopene Is Enriched in Tomato Fruit by CRISPR/Cas9-Mediated Multiplex Genome Editing. Front. Plant Sci. 2018, 9, 559.
- Nakayasu, M.; Akiyama, R.; Lee, H.J.; Osakabe, K.; Osakabe, Y.; Watanabe, B.; Sugimoto, Y.; Umemoto, N.; Saito, K.; Muranaka, T.; et al. Generation of α-solanine-free hairy roots of potato by CRISPR/Cas9 mediated genome editing of the St16DOX gene. Plant Physiol. Biochem. 2018, 131, 70–77.
- Ito, Y.; Nishizawa-Yokoi, A.; Endo, M.; Mikami, M.; Toki, S. CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 2015, 467, 76–82.
- Lang, Z.; Wang, Y.; Tang, K.; Tang, D.; Datsenka, T.; Cheng, J.; Zhang, Y.; Handa, A.K.; Zhu, J.K. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. USA 2017, 114, E4511–E4519.
- Wang, D.; Samsulrizal, A.; Yan, B.C.; Allcock, C.; Craigon, D.J. Characterization of CRISPR Mutants Targeting Genes Modulating Pectin Degradation in Ripening Tomato. Plant Physiol. 2019, 179, 544–557.
- Yu, Q.H.; Wang, B.; Li, N.; Tang, Y.; Yang, S.; Yang, T.; Xu, J.; Guo, C.; Yan, P.; Wang, Q.; et al. CRISPR/Cas9-induced Targeted Mutagenesis and Gene Replacement to Generate Long-shelf Life Tomato Lines. Sci. Rep. 2017, 7, 11874.




