
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Benyamin Rosental | + 3140 word(s) | 3140 | 2021-07-29 10:57:36 | | | |
| 2 | Conner Chen | -35 word(s) | 3105 | 2021-08-04 11:25:21 | | |
Video Upload Options
The immune system has evolved to protect organisms from infections caused by bacteria, viruses, and parasitic pathogens. In addition, it provides regenerative capacities, tissue maintenance, and self/non-self recognition of foreign tissues. In general, innate immunity is a fast and non-specific response associated with the presence of humoral and cellular elements. By contrast, adaptive immunity uses the induction of specialized cells such as B and T lymphocytes and molecules including the major histocompatibility complex (MHC), B-cell receptors (BCR), T-cell receptors (TCR), immunoglobulins (Ig), and antibodies to confer immunological memory and very high specificity thus fighting against a previously recognized infection. Both kinds of immune responses rely on two main cellular activities which are phagocytosis and cytotoxicity. These cellular immune mechanisms have been found at the earliest evolutionary stages of multicellular animals and diversified into a wide heterogeneous repertoire of effector cells through evolution.
1. Phagocytosis in the Evolution of Immune Response
Phagocytosis is the process by which particles (>0.5 μm) are recognized, bound to a plasma-membrane envelope, and internalized into an organelle called “phagosome”. Two major classes of particles are engulfed by phagocytes: foreign microorganisms and “altered self” (apoptotic and necrotic cells) particles. Élie Metchnikoff was the first to realize the significance of phagocytosis for cellular immunity in processes such as the host response to injury and infection, inflammation, and tissue homeostasis [1][2][3]. Phagocytosis is highly conserved between species and has essential roles for the development and maintenance of multicellular organisms. The phagocytic process requires a coordinated sequence of events that begins with the engagement of plasma-membrane receptors, which allow the recognition of damaged or infected cells and different classes of pathogens. This leads to the activation of signaling pathways needed for the rearrangement of the cytoskeleton and the internalization of the particle. Once internalized, the phagosome dynamically undergoes fusion and fission events with intracellular secretory vesicles to finally mature into a phagolysosome. The hydrolytic activity of phagolysosomes results in the digestion of the internalized cargo [1].
1.1. Phagocytic Effector Cells
Amebocytes are the most primitive animal cells with phagocytic ability, these are found in sponges and cnidarians acting in the recognition of nutrients and foreign elements [4]. We have recently characterized two populations of phagocytes (granular spheroid and ameboid phenotype) from members of the Hexacorallia class, the coral Pocillopora damicornis and the sea anemone Nematostella vectensis, finding that they engulf bacteria, fungal antigens, carboxylated beads and self-damaged cells. We showed that this phagocytosis is different than pinocytosis which is performed by the majority of the cells [5]. Reticular cells, the phagocytic cells of free living planarians from the phylum Platyhelminthes, can migrate towards heat-shocked bacteria and phagocytose them. Their pseudopodia and the modes of their endocytosis can be morphologically compared to human neutrophils [6][7]. From the phylum Annelida, the earthworm Eisenia andrei has several subtypes of coelomocytes including eleocytes and granular amoebocytes [8]. In arthropods, several cell types have been characterized. For example, Drosophila melanogaster has hemolymph circulating cells collectively called hemocytes. Two types of hemocytes have been identified in the larval stage: Plasmatocytes, which phagocytose microorganisms and cell debris and secrete signaling molecules (i.e., Eiger, Upds, drosocin, defensins) with high similarity to mammalian macrophages and neutrophils, according to transcriptional profiles; and crystal cells, which activate the melanization cascade upon wounds or infections [9]. However, single-cell RNA sequencing studies showed that these cell types are more heterogeneous as 14 different functional clusters of hematocytes have been recently discovered, including lamellocytes, which are activated immune cells that only differentiate upon immune induction [10]. The tunicate Botryllus schlosseri has a myeloid lineage of phagocytic cells with high similarities and gene set homology to mammalian myeloid lineage [11], and also amoebocytes and large phagocytes suggested to be related to arthropods and echinoderms [12]. In bony fishes, the phagocytic armament includes macrophages, monocytes, dendritic cells, neutrophils, granulocytes, eosinophils, basophils and mast cells [13]. While in mammals, these professional phagocytes are able to differentiate into highly specialized subtypes of cells which exert different cytokine profiles, exclusive functions in regeneration and infection fighting (i.e., M1 and M2 macrophages), tissue specificity and also novel molecular mechanisms such as the antibody-dependent cellular phagocytosis (ADCP) for the clearance of microorganisms and immune complexes [14][15].
1.2. Receptors
Phagocytic cell lineages possess cell-surface receptors as the mechanism to respond and initiate the phagocytic process and the production of killing molecules. Primitive sponges possess LPS binding receptors and intracellular receptors “Nucleotide-binding domain and Leucine-rich Repeat” (NLRs) to bind fungal polysaccharides, bacteria and virus [16][17]. Toll-like receptors (TLRs) are ancient sensors and the best characterized in detecting and respond against invading pathogens. TLRs have been traced to several invertebrate species as their emergence predate the separation of bilaterians and cnidarians. In D. melanogaster, Toll receptors have a crucial role in immune defense. Flies deficient in Toll members are severely immunocompromised to response against fungal and bacterial infections [18]. Moreover, the Down syndrome cell-adhesion molecule (DsCAM) acts as a phagocytic cell-surface receptor that binds to bacteria such as Escherichia coli [19]. A new study in B. schlosseri has identified BsTLR1 as a member of the TLR family which is actively transcribed in phagocytes and morula cells as a mechanism of non-self recognition [20]. Interestingly, TLRs are highly diversified in urchins (253 genome sequences), more than in vertebrates [21]. Agnathans have seven identified TLRs, while in mammals this number is up to 13, which have high specificity in the ligands they recognize, suggesting a host–ligand coevolution [22]. Although TLRs have not been identified in planarians, extracellular leucine rich repeat (LRR)-motifs are coded in their genome [23]; however, the exact role LRRs play in the immune response is unknow. Other types of receptors are highly conserved through evolution. For example, the scavenger receptors Croquemort (Crq) and Peste of D. melanogaster have convergently evolved with their mammalian orthologues CD36 and SR-BI, respectively [24]. There are also three predicted CD36-family homologs in the genome of Caenorhabditis elegans [25]. They are CD36-like family members and major plasmatocyte markers that recognize apoptotic cells and bind to surface lipopetides of different Gram-negative and Gram-positive bacteria and fungi. Both are required to efficiently eliminate infections, and their absence causes poor phagocytic plasmatocytes and defects on phagosome maturation [26]. The Transformer (Trf) proteins also have important roles in the immunity of sea urchins by inducing the phagocytic activity of coelomocytes to directly engulf bacteria in an ADCP resembling matter [27][28]. SpTrf genes have two exons, recombination mechanisms between the gene elements of the second exon result in the fact that sea urchins express more than 260 different Trfs proteins, single one recombinant protein expressed in an individual cell, which act synergistically to detect a variety of pathogens thus providing an efficient immunological army [29][30]. Furthermore, no Trfs have been identified outside the echinoid lineage [31], indicating that there are several unique immune mechanisms that should be studied in sea urchins. Moreover, this discovery suggests that there are diverse mechanisms of immune receptor diversification, which are different from what we know in vertebrates.
2. Cellular Cytotoxicity in the Evolution of Immune Response against Abnormal Self and Pathogens
Cytotoxicity (cell-mediated killing) is the process in which certain cells secrete “toxins” that lysate and neutralize unwanted target cells (i.e., pathogens, viral-infected cells, tumoral cells, foreign transplanted cells). Cooper has defined that through evolution, immune-mediated cytotoxicity could be classified into level I and level II cytotoxicity. Level I in mammals is mediated by macrophages and neutrophils which release toxic granules in the inflammatory area. They are considered the most basic manifestations of cytotoxicity and are present in invertebrates (sponges, coelenterates, sipunculids, annelids, mollusks, arthropods, echinoderms and protochordates). In contrast, level II or acquired cytotoxicity is mediated by cells that require a specific induction to be activated, through specific recognition, such as allogeneic responses, or induced by adaptive molecules. These include cytotoxic T lymphocytes (CTL), natural killer (NK), and antibody-dependent cell-mediated cytotoxicity (ADCC). It seems that these cells appear later in evolution [32][33], but the characterization of specific allogeneic responses suggests that they might be in more animal groups than what is currently known. Since the work of Cooper in 1980, NK cells were shown to have specificity and recognition of targets for cytotoxicity trough immunological synapse without the killing of adjacent cells [34][35], for that reason we moved NK-like recognition based cytotoxicity mechanism to level II cytotoxicity, compared to the original definition by Cooper [33].
All cytotoxic cells share certain characteristics such as (1) the ability to trigger cell death of target cells, (2) the presence of adhesion molecules allowing their adhesion to the target, (3) their activity is mediated by monokines and lymphokines [36].
2.1. Effector Cells
All animals have several immune cells with cytotoxic activity to lyse host and foreign cells. Regarding level I cytotoxicity, those cells have been found in different invertebrate organisms as probably the most ancient type of cytotoxic immune response. For example, killer cells from sipunculid worms exert cytotoxic effects on allogeneic erythrocytes, but not on erythrocytes from worms that reside closely [37]. In the freshwater pulmonated mollusk Planorbarius corneus, a class of hemocyte that has been morphologically characterized as round hemocytes (RH) exerts cytotoxic activity on the human erythroleukemia K562 cells thus participating in the graft rejection of allo- and xeno-grafts [36]. In arthropods, the amoeboid hemocytes of Limolus polyphemus are stimulated by bacterial lipopolysaccharide (LPS) endotoxins to secrete great amounts of clottable protein and to produce nitric oxide (NO) synthesis as a cytotoxic mechanism to protect the host from invading pathogens [38]. Additionally, in in vitro experiments, crayfish (Astacus astacus) granular and semi granular hemocytes with phenoloxidase and laccase activity display cytotoxic effects towards different mammalian tumor and non-tumor cells [39][40]. Coelomocytes from the sea urchin Arbacia punctulata exert cytotoxic activity against human and murine target cells. A particular population of these phagocytic cells is the one with the highest cytotoxic activity which are positive to the human NK markers CD14, CD56 and CD158b [41]. These cytotoxic cells have also found in Strongylocentrotus droebachiensis, S. pallidus and Echinus esculentus in which targeted-cell death is characterized by cell detachment, disintegration and the formation of a multinuclear non-cellular protoplasm [42].
At this point we do not think that phagocytosis of damaged self-cells should be considered as level 1 cytotoxicity. Although, we do think that where phagocytosis of damaged self-cells is found, it can be suggestive of potential level 1 cytotoxicity (as the mechanisms of altered-self recognition exist) and should be further investigated (Figure 1). For instance, the recent discovery of phagocytosis of heat-stressed cells in ex-vivo experiments in Hexacorallians [5].
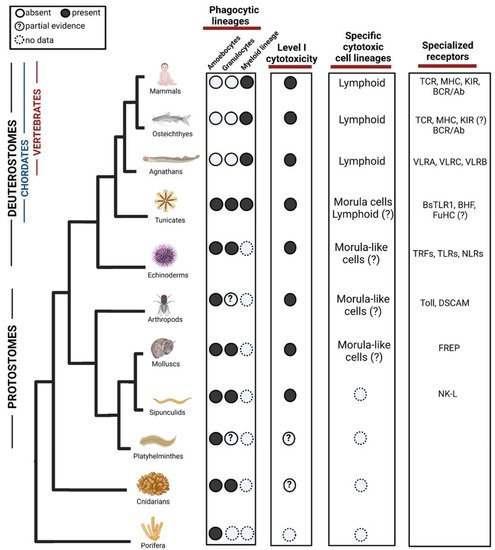
Figure 1. Evolution of phagocytic and cytotoxic cell lineages in the immune system. Among phagocytic lineages, amoebocytes and granulocytes are present in invertebrates, while myeloid lineage seems to evolve in the ancestor of tunicates and vertebrates. Cells representing the Level I cytotoxicity exist in Protostomes but might appear in cnidarian. Among specific cytotoxic cell lineages, morula cells have been found in tunicates although partial evidence suggests that they may evolve in earlier ancestors. Lymphoid cells are found in agnathans and have diversified in vertebrates, there are suggestions of their form in Tunicates but not validated yet. In the third column, we show examples of diversification of specialized receptors acting in both phagocytosis and cytotoxicity. TLR = Toll-like receptors, TCR = T-cell receptor, MHC = Major histocompatibility complex, KIR = Killer Inhibitory Receptors, BCR = B cell receptor, VLR = Variable lymphocyte receptor, BHF = Botryllus histocompatibility factor, TRF = Transformer proteins, NLR = NOD like receptors, DSCAM = Down syndrome cell adhesion molecule, FREP = Fibrinogen-related proteins, NK-L = Natural killer-like receptor. ?—Represents missing functional or molecular validation. Created with BioRender.com. Data sources are summarized in Table 1.
Table 1. Data sources for Figure 1.
| Animal Group | Effector Cells | Receptors and Effector Molecules |
|---|---|---|
| Mammals | (Wolff and Humeniuk 2013) [43] | (Vandendriessche, Cambier et al., 2021) [14] |
| (Rosental, Appel et al., 2012) [35] | ||
| (Hirayama, Iida et al., 2017) [15] | ||
| (Geffner 2005) [44] | ||
| Osteichthyes | (Potts and Bowman 2017) [45] | (Wei, Zhou et al., 2007) [46] |
| (Moss, Monette et al., 2009) [47] | (Kasahara and Flajnik 2019) [48] | |
| (Tang, Iyer et al., 2017) [49] | (Flajnik and Du Pasquier 2004) [50] | |
| (Flajnik 2018) [51] | ||
| Agnathans | (Han, Das et al., 2015) [52] | (Pancer, Amemiya et al., 2004) [53] |
| (Hirano, Guo et al., 2013) [54] | ||
| (Das, Li et al., 2015) [55] | ||
| (Mayer, Uinuk-ool et al., 2002) [56] | ||
| Tunicates | (Rosental, Kowarsky et al., 2018) [11] | (Voskoboynik, Newman et al., 2013) [57] |
| (Rosental, Raveh et al., 2020) [12] | (Nyholm, Passegue et al., 2006) [58] | |
| (Peronato, Franchi et al., 2020) [20] | ||
| Echinoderms | (Arizza, Giaramita et al., 2007) [59] | (Yakovenko, Donnyo et al., 2021) [27] |
| (Cooper 1980) [33] | (Chou, Lun et al., 2018) [28] | |
| (Lin, Zhang et al., 2001) [41] | ||
| Arthropods | (Muñoz-Chápuli, Carmona et al., 2005) [60] | (Melcarne, Lemaitre et al., 2019) [61] |
| (Lanot, Zachary et al., 2001) [62] | ||
| (Meister and Lagueux 2003) [63] | ||
| (Cattenoz, Sakr et al., 2020) [10] | ||
| (Csordás, Gábor et al., 2021) [9] | ||
| (Cárdenas, Dankert et al., 2004) [64] | ||
| Molluscs | (Franceschi, Cossarizza et al., 1991) [36] | (Schultz, Bu et al., 2018) [65] |
| (Nakayama, Nomoto et al., 1997) [66] | (Loker, Adema et al., 2004) [67] | |
| Sipunculids | (Boiledieu and Valembois 1977) [37] | (Flajnik and Du Pasquier 2004) [50] |
| Platyelminthes | (de Oliveira, Lopes et al., 2018) [68] | (Peiris, Hoyer et al., 2014) [23] |
| (Morita 1991) [6] | ||
| Cnidarians | (Snyder G 2021) [5] | |
| Porifera | (Mukherjee, Ray et al., 2015) [4] | (Pita, Hoeppner et al., 2018) [69] |
| (Yuen, Bayes et al., 2014) [16] | ||
| (Wiens, Korzhev et al., 2005) [17] |
For level II of specific cytotoxicity, in the tunicate B. schlosseri, morula cells (MC) have been characterized as cytotoxic cells. They contain phenoloxidase, accumulate in rejection points and morphologically resembles NK cells. While 15% of their genes are shared with cytotoxic lymphocytes of vertebrates, MC express 85% tunicate-specific gene repertoire [11][12], suggestive of convergent lineage more resembling other invertebrates phenoloxidase based immune cells (Figure 1). The cytotoxic activity of MCs is involved in a natural process of allorecognition that occurs when two B. schlosseri colonies with different genotypes touch [70]. This self–nonself recognition is regulated by a single polymorphic histocompatibility gene: the Botryllus Histocompatibility Factor (BHF) [57]. The sharing of at least one BHF allele is a determinant for the fusion or rejection of the colonies [11][12]. Interestingly, MC express high levels of the polymorphic gene FuHC which could be a base for BHF recognition mechanism [12][71] (Figure 1).
Transcriptome analysis and cellular characterization studies in the jawless vertebrates, lampreys, have proposed that two T-like cell populations expressing the variable lymphocyte receptors (VLRs) VLRA and VLRC, respectively, carries cytotoxic activity [55][56]. These cells express several genes including cell surface receptors (i.e., CD45, TCR, VpreB, paired-Ig-like receptors), cytokines (IL17) and transcription factors (i.e., GATA 2/3, c-Rel, BCL11b) resembling those that T lymphocytes use to migrate, proliferate and differentiate in jawed vertebrates [72]. Lymphocyte lineages have also been found in the hagfish, however these have not been fully characterized [55][73]. This shows a whole convergent parallel adaptive immune system of lymphocytes, which is based on LLRs instead of Ig superfamily like the jawed vertebrates (Figure 1).
In jawed vertebrates, the cytotoxic cells NK and CTLs belong to the lymphoid lineage. NK cells do not express antigen-specific receptors but are highly capable to recognize and kill tumor and viral-infected cells [74]. NK cell function has been recognized in representatives of all vertebrate classes [51]. Comparative studies using high-quality genome assemblies have shown that NK cells diversity in mammals has a big influence on species-specific immune responses and outcomes of pathogen infection [75]. NKT-like cells have been identified in fishes [51][76] the frog Xenopus laevis [77] and through the identification of CD1 molecules, it is suggested that this kind of cells are also present in reptiles and birds [51].
CTLs have three important elements to recognize antigens expressed by viral-infected cells or cancer tissue: (1) a diverse repertoire of Ig domain-based receptors, (2) T cell receptor (TCR) and (3) major histocompatibility complex (MHC). Genes of these elements and the presence of CTLs is a common feature found in all jawed vertebrates which include cartilaginous and bony fishes, amphibians, reptiles, birds, and mammals. Moreover, these cells were also found in the extinct placoderms [51][78]. In mammals, also activated macrophages could selectively lyse malignant cells in a contact-dependent and non-phagocytic process, having important roles in the restoration of tissue homeostasis [79]. Finally, ADCC is well characterized in mammals by eosinophilic and neutrophilic granulocytes, NK cells, and CD16+ blood monocytes [44]. Their main effects are associated with graft rejection, autoimmune diseases, tumor surveillance, antiviral and antiparasitic defense [44][80][81][82].
We think that in cases where allogeneic specificity is found in cytotoxicity, this should be considered and further investigated as potential for level II cytotoxicity. For instance, the example mentioned earlier of sipunculid worms showing different cytotoxic effects between levels of allogeneic proximity of the target erythrocytes is suggestive of specificity recognition by the cytotoxic effector cells [37]. Characterization of the cells and those recognition mechanisms would shed light if there were a presence of level II specific cytotoxicity. Moreover, receptor recombination as an example of Trfs in urchins could also suggest specific cytotoxicity, at this stage it was only tested against bacterial opsonization [27][28], but if it is effective against foreign cells or abnormal self, then could be considered as level II specific lysis.
2.2. Receptors
Killer cells possess diverse receptors for triggering their cytotoxic functions, these receptors seem to appear early in evolution although without high specificity. In mollusks, the Fibrinogen-related proteins (FREPs) bind to parasites and has been suggested as potential adaptive defense molecules [50][65][67]. Hemocyte membranes of the arthropod P. bicarinatus have trypsin-labile receptors and nonspecific cytotoxic cell receptors protein 1 (NCCRP-1) which appear to be mediators of allorecognition [64]. Additionally, haemocytes from Prodenia eridania express non-specific receptors for foreign particles [33]. NK cells from mammals have Killer Inhibitory Receptors (KIRs) which inhibit the activation of cytotoxic programs when recognize self-MHC, according to the “missing self” hypothesis NK cells kill in this manner allogeneic foreign cells that do not express self-MHC or that have altered expression of MHC molecules [83][84][85]. In humans, CD94, NKG2 and NKR-P1 are examples of these receptors, while rodents express a group of Ly49 receptors. Blood cells of the tunicate B. schlosseri also express a gene coding for a type II transmembrane protein with high similarity to vertebrates CD94 and NKR-P1, which is upregulated during allorecognition [86]. VLRA, VLRB and VLRC genes have been identified in lampreys and hagfish [55][56]. These receptors are highly diverse due to a somatic diversification strategy based on the insertion of LRR sequences in a single germline gene [53]. The locus VLRA has >500 LRR cassettes, while there are >800 LRR cassettes in the VLRB locus and >200 LRR cassettes in the VLRC locus [87][88]. This evolutionary mechanism to diversify lymphocyte receptors at a single cell level is similar, but not identical, to the multigene recombinatorial strategy of immunoglobulin gene segments and TCR molecules in jawed vertebrates [53][89].
In all jawed vertebrates, CTLs recognize peptides bound to MHC molecules by expressing the T cell receptor complex (TCR). Peptides are derived from viral or bacterial proteins during infection [90], and MHC I nonself recognition also occurs after allotransplantation [91]. In this sense, the expression of TCRs is clonotypic since one T cell expresses a single TCR specificity that is determined by both the antigen peptide and the MHC determinants. TCR diversity, which is generated by mechanisms of genomic rearrangement, and the fact that MHC molecules are highly polymorphic allow organisms to counteract pathogen evasion of T cell responses [92]. While TCR is the major CTL marker, the glycoproteins CD3, CD8 and CD57 are other important markers which bind MHC molecules [92]. The structure and function of immunoglobulin M (IgM), the classic TCRs (αβ chains) and MHC class II are conserved in jawed vertebrates from cartilaginous and bony fishes, to amphibians, reptiles, birds, and mammals. However, other immune receptors as γδ TCR, NKRs and nonclassical MHCs are present but with diverse function, which in most cases is still unexplored [51][48][93][94].
References
- Gordon, S. Phagocytosis: An Immunobiologic Process. Immunity 2016, 44, 463–475.
- Gordon, S. Elie Metchnikoff, the Man and the Myth. J. Innate Immun. 2016, 8, 223–227.
- Metschnikoff, E. Memoirs: The Ancestral History of the Inflammatory Process. J. Cell Sci. 1884, s2–s24, 112–117.
- Mukherjee, S.; Ray, M.; Ray, S. Phagocytic efficiency and cytotoxic responses of Indian freshwater sponge (Eunapius carteri) cells isolated by density gradient centrifugation and flow cytometry: A morphofunctional analysis. Zoology 2015, 118, 8–18.
- Snyder, G.A.; Eliachar, S.; Connelly, M.T.; Talice, S.; Hadad, U.; Gershoni-Yahalom, O.; Browne, W.E.; Palmer, C.V.; Rosental, B.; Traylor-Knowles, N. Functional characterization of Hexacorallia phagocytic cells. Front. Immunol. 2021, 12, 2402.
- Morita, M. Phagocytic response of planarian reticular cells to heat-killed bacteria. Hydrobiologia 1991, 227, 193–199.
- Maciel, E.I.; Oviedo, N.J. Platyhelminthes: Molecular Dissection of the Planarian Innate Immune System. In Advances in Comparative Immunology; Cooper, E.L., Ed.; Springer: Cham, Switzerland, 2018.
- Homa, J. Earthworm coelomocyte extracellular traps: Structural and functional similarities with neutrophil NETs. Cell Tissue Res. 2018, 371, 407–414.
- Csordás, G.; Gábor, E.; Honti, V. There and back again: The mechanisms of differentiation and transdifferentiation in Drosophila blood cells. Dev. Biol. 2021, 469, 135–143.
- Cattenoz, P.B.; Sakr, R.; Pavlidaki, A.; Delaporte, C.; Riba, A.; Molina, N.; Hariharan, N.; Mukherjee, T.; Giangrande, A. Temporal specificity and heterogeneity of Drosophila immune cells. EMBO J. 2020, 39, e104486.
- Rosental, B.; Kowarsky, M.; Seita, J.; Corey, D.M.; Ishizuka, K.J.; Palmeri, K.J.; Chen, S.-Y.; Sinha, R.; Okamoto, J.; Mantalas, G.; et al. Complex mammalian-like haematopoietic system found in a colonial chordate. Nature 2018, 564, 425–429.
- Rosental, B.; Raveh, T.; Voskoboynik, A.; Weissman, I.L. Evolutionary perspective on the hematopoietic system through a colonial chordate: Allogeneic immunity and hematopoiesis. Curr. Opin. Immunol. 2020, 62, 91–98.
- Grayfer, L.; Kerimoglu, B.; Yaparla, A.; Hodgkinson, J.W.; Xie, J.; Belosevic, M. Mechanisms of Fish Macrophage Antimicrobial Immunity. Front. Immunol. 2018, 9, 1105.
- Vandendriessche, S.; Cambier, S.; Proost, P.; Marques, P.E. Complement Receptors and Their Role in Leukocyte Recruitment and Phagocytosis. Front. Cell Dev. Biol. 2021, 9, 624025.
- Hirayama, D.; Iida, T.; Nakase, H. The Phagocytic Function of Macrophage-Enforcing Innate Immunity and Tissue Homeostasis. Int. J. Mol. Sci. 2017, 1, 92.
- Yuen, B.; Bayes, J.M.; Degnan, S.M. The characterization of sponge NLRs provides insight into the origin and evolution of this innate immune gene family in animals. Mol. Biol. Evol. 2014, 31, 106–120.
- Wiens, M.; Korzhev, M.; Krasko, A.; Thakur, N.L.; Perović-Ottstadt, S.; Breter, H.J.; Ushijima, H.; Diehl-Seifert, B.; Müller, I.M.; Müller, W.E. Innate immune defense of the sponge Suberites domuncula against bacteria involves a MyD88-dependent signaling pathway. Induction of a perforin-like molecule. J. Biol. Chem. 2005, 280, 27949–27959.
- Narbonne-Reveau, K.; Charroux, B.; Royet, J. Lack of an antibacterial response defect in Drosophila Toll-9 mutant. PLoS ONE 2011, 6, e17470.
- Stuart, L.M.; Ezekowitz, R.A. Phagocytosis and comparative innate immunity: Learning on the fly. Nat. Rev. Immunol. 2008, 8, 131–141.
- Peronato, A.; Franchi, N.; Loriano, B. BsTLR1: A new member of the TLR family of recognition proteins from the colonial ascidian Botryllus schlosseri. Fish Shellfish Immunol. 2020, 106, 967–974.
- Buckley, K.M.; Rast, J.P. Dynamic evolution of toll-like receptor multigene families in echinoderms. Front. Immunol. 2012, 3, 136.
- Liu, G.; Zhang, H.; Zhao, C.; Zhang, H. Evolutionary History of the Toll-Like Receptor Gene Family across Vertebrates. Genome Biol. Evol. 2020, 12, 3615–3634.
- Peiris, T.H.; Hoyer, K.K.; Oviedo, N.J. Innate immune system and tissue regeneration in planarians: An area ripe for exploration. Semin. Immunol. 2014, 26, 295–302.
- Guillou, A.; Troha, K.; Wang, H.; Franc, N.C.; Buchon, N. The Drosophila CD36 Homologue croquemort is required to Maintain Immune and Gut Homeostasis during Development and Aging. PLoS Pathog. 2016, 12, e1005961.
- Means, T.K.; Mylonakis, E.; Tampakakis, E.; Colvin, R.A.; Seung, E.; Puckett, L.; Tai, M.F.; Stewart, C.R.; Pukkila-Worley, R.; Hickman, S.E.; et al. Evolutionarily conserved recognition and innate immunity to fungal pathogens by the scavenger receptors SCARF1 and CD36. J. Exp. Med. 2009, 206, 637–653.
- Febbraio, M.; Hajjar, D.P.; Silverstein, R.L. CD36: A class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J. Clin. Investig. 2001, 108, 785–791.
- Yakovenko, I.; Donnyo, A.; Ioscovich, O.; Rosental, B.; Oren, M. The Diverse Transformer (Trf) Protein Family in the Sea Urchin Paracentrotus lividus Acts through a Collaboration between Cellular and Humoral Immune Effector Arms. Int. J. Mol. Sci. 2021, 22, 6639.
- Chou, H.-Y.; Lun, C.M.; Smith, L.C. SpTransformer proteins from the purple sea urchin opsonize bacteria, augment phagocytosis, and retard bacterial growth. PLoS ONE 2018, 13, e0196890.
- Oren, M.; Barela Hudgell, M.A.; D’Allura, B.; Agronin, J.; Gross, A.; Podini, D.; Smith, L.C. Short tandem repeats, segmental duplications, gene deletion, and genomic instability in a rapidly diversified immune gene family. BMC Genom. 2016, 17, 900.
- Smith, L.C.; Lun, C.M. The SpTransformer Gene Family (Formerly Sp185/333) in the Purple Sea Urchin and the Functional Diversity of the Anti-Pathogen rSpTransformer-E1 Protein. Front. Immunol. 2017, 8, 725.
- Majeske, A.J.; Oren, M.; Sacchi, S.; Smith, L.C. Single sea urchin phagocytes express messages of a single sequence from the diverse Sp185/333 gene family in response to bacterial challenge. J. Immunol. 2014, 193, 5678–5688.
- Buchmann, K. Evolution of Innate Immunity: Clues from Invertebrates via Fish to Mammals. Front. Immunol. 2014, 5, 459.
- Cooper, E.L. Phylogeny of cytotoxicity. Endeavour 1980, 4, 160–165.
- Colantonio, A.D.; Bimber, B.N.; Neidermyer, W.J., Jr.; Reeves, R.K.; Alter, G.; Altfeld, M.; Johnson, R.P.; Carrington, M.; O’Connor, D.H.; Evans, D.T. KIR polymorphisms modulate peptide-dependent binding to an MHC class I ligand with a Bw6 motif. PLoS Pathog. 2011, 7, e1001316.
- Rosental, B.; Appel, M.Y.; Yossef, R.; Hadad, U.; Brusilovsky, M.; Porgador, A. The effect of chemotherapy/radiotherapy on cancerous pattern recognition by NK cells. Curr. Med. Chem. 2012, 19, 1780–1791.
- Franceschi, C.; Cossarizza, A.; Ortolani, C.; Monti, D.; Ottaviani, E. Natural cytotoxicity in a freshwater pulmonate mollusc: An unorthodox comparative approach. Adv. Neuroimmunol. 1991, 1, 99–113.
- Boiledieu, D.; Valembois, P. Natural cytotoxic activity of sipunculid leukocytes on allogenic and xenogenic erythrocytes. Dev. Comp. Immunol. 1977, 1, 207–216.
- Radomski, M.W.; Martin, J.F.; Moncada, S. Synthesis of Nitric Oxide by the Haemocytes of the American Horseshoe Crab (Limulus polyphemus). Philos. Trans. Biol. Sci. 1991, 334, 129–133.
- Söderhäll, K.; Wingren, A.; Johansson, M.W.; Bertheussen, K. The cytotoxic reaction of hemocytes from the freshwater crayfish, Astacus astacus. Cell. Immunol. 1985, 94, 326–332.
- Cárdenas, W.; Dankert, J.R. Cresolase, catecholase and laccase activities in haemocytes of the red swamp crayfish. Fish Shellfish Immunol. 2000, 10, 33–46.
- Lin, W.; Zhang, H.; Beck, G. Phylogeny of natural cytotoxicity: Cytotoxic activity of coelomocytes of the purple sea urchin, Arbacia punctulata. J. Exp. Zool. 2001, 290, 741–750.
- Gross, P.S.; Al-Sharif, W.Z.; Clow, L.A.; Smith, L.C. Echinoderm immunity and the evolution of the complement system. Dev. Comp. Immunol. 1999, 23, 429–442.
- Wolff, L.; Humeniuk, R. Concise review: Erythroid versus myeloid lineage commitment: Regulating the master regulators. Stem cells 2013, 31, 1237–1244.
- Geffner, J. Antibody-Dependent Cellular Cytotoxicity. In Encyclopedia of Immunotoxicology; Vohr, H.-W., Ed.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 1–5.
- Potts, K.S.; Bowman, T.V. Modeling Myeloid Malignancies Using Zebrafish. Front. Oncol. 2017, 7, 297.
- Wei, S.; Zhou, J.M.; Chen, X.; Shah, R.N.; Liu, J.; Orcutt, T.M.; Traver, D.; Djeu, J.Y.; Litman, G.W.; Yoder, J.A. The zebrafish activating immune receptor Nitr9 signals via Dap12. Immunogenetics 2007, 59, 813–821.
- Moss, L.D.; Monette, M.M.; Jaso-Friedmann, L.; Leary, J.H., 3rd; Dougan, S.T.; Krunkosky, T.; Evans, D.L. Identification of phagocytic cells, NK-like cytotoxic cell activity and the production of cellular exudates in the coelomic cavity of adult zebrafish. Dev. Comp. Immunol. 2009, 33, 1077–1087.
- Kasahara, M.; Flajnik, M.F. Origin and evolution of the specialized forms of proteasomes involved in antigen presentation. Immunogenetics 2019, 71, 251–261.
- Tang, Q.; Iyer, S.; Lobbardi, R.; Moore, J.C.; Chen, H.; Lareau, C.; Hebert, C.; Shaw, M.L.; Neftel, C.; Suva, M.L.; et al. Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing. J. Exp. Med. 2017, 214, 2875–2887.
- Flajnik, M.F.; Du Pasquier, L. Evolution of innate and adaptive immunity: Can we draw a line? Trends Immunol. 2004, 25, 640–644.
- Flajnik, M.F. A cold-blooded view of adaptive immunity. Nat. Rev. Immunol. 2018, 18, 438–453.
- Han, Q.; Das, S.; Hirano, M.; Holland, S.J.; McCurley, N.; Guo, P.; Rosenberg, C.S.; Boehm, T.; Cooper, M.D. Characterization of Lamprey IL-17 Family Members and Their Receptors. J. Immunol. 2015, 195, 5440–5451.
- Pancer, Z.; Amemiya, C.T.; Ehrhardt, G.R.A.; Ceitlin, J.; Larry Gartland, G.; Cooper, M.D. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 2004, 430, 174–180.
- Hirano, M.; Guo, P.; McCurley, N.; Schorpp, M.; Das, S.; Boehm, T.; Cooper, M.D. Evolutionary implications of a third lymphocyte lineage in lampreys. Nature 2013, 501, 435–438.
- Das, S.; Li, J.; Hirano, M.; Sutoh, Y.; Herrin, B.R.; Cooper, M.D. Evolution of two prototypic T cell lineages. Cell. Immunol. 2015, 296, 87–94.
- Mayer, W.E.; Uinuk-ool, T.; Tichy, H.; Gartland, L.A.; Klein, J.; Cooper, M.D. Isolation and characterization of lymphocyte-like cells from a lamprey. Proc. Natl. Acad. Sci. USA 2002, 99, 14350–14355.
- Voskoboynik, A.; Newman, A.M.; Corey, D.M.; Sahoo, D.; Pushkarev, D.; Neff, N.F.; Passarelli, B.; Koh, W.; Ishizuka, K.J.; Palmeri, K.J.; et al. Identification of a colonial chordate histocompatibility gene. Science 2013, 341, 384–387.
- Nyholm, S.V.; Passegue, E.; Ludington, W.B.; Voskoboynik, A.; Mitchel, K.; Weissman, I.L.; De Tomaso, A.W. Fester, a candidate allorecognition receptor from a primitive chordate. Immunity 2006, 25, 163–173.
- Arizza, V.; Giaramita, F.T.; Parrinello, D.; Cammarata, M.; Parrinello, N. Cell cooperation in coelomocyte cytotoxic activity of Paracentrotus lividus coelomocytes. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007, 147, 389–394.
- Muñoz-Chápuli, R.; Carmona, R.; Guadix, J.A.; Macías, D.; Pérez-Pomares, J.M. The origin of the endothelial cells: An evo-devo approach for the invertebrate/vertebrate transition of the circulatory system. Evol. Dev. 2005, 7, 351–358.
- Melcarne, C.; Lemaitre, B.; Kurant, E. Phagocytosis in Drosophila: From molecules and cellular machinery to physiology. Insect Biochem. Mol. Biol. 2019, 109, 1–12.
- Lanot, R.; Zachary, D.; Holder, F.; Meister, M. Postembryonic Hematopoiesis in Drosophila. Dev. Biol. 2001, 230, 243–257.
- Meister, M.; Lagueux, M. Drosophila blood cells. Cell. Microbiol. 2003, 5, 573–580.
- Cárdenas, W.; Dankert, J.R.; Jenkins, J.A. Flow cytometric analysis of crayfish haemocytes activated by lipopolysaccharides. Fish Shellfish Immunol. 2004, 17, 223–233.
- Schultz, J.H.; Bu, L.; Adema, C.M. Comparative immunological study of the snail Physella acuta (Hygrophila, Pulmonata) reveals shared and unique aspects of gastropod immunobiology. Mol. Immunol. 2018, 101, 108–119.
- Nakayama, K.; Nomoto, A.M.; Nishijima, M.; Maruyama, T. Morphological and Functional Characterization of Hemocytes in the Giant ClamTridacna crocea. J. Invertebr. Pathol. 1997, 69, 105–111.
- Loker, E.S.; Adema, C.M.; Zhang, S.M.; Kepler, T.B. Invertebrate immune systems--not homogeneous, not simple, not well understood. Immunol. Rev. 2004, 198, 10–24.
- De Oliveira, M.S.; Lopes, K.A.R.; Leite, P.M.S.C.M.; Morais, F.V.; de Campos Velho, N.M.R. Physiological evaluation of the behavior and epidermis of freshwater planarians (Girardia tigrina and Girardia sp.) exposed to stressors. Biol. Open 2018, 7, 6.
- Pita, L.; Hoeppner, M.P.; Ribes, M.; Hentschel, U. Differential expression of immune receptors in two marine sponges upon exposure to microbial-associated molecular patterns. Sci. Rep. 2018, 8, 16081.
- Corey, D.M.; Rosental, B.; Kowarsky, M.; Sinha, R.; Ishizuka, K.J.; Palmeri, K.J.; Quake, S.R.; Voskoboynik, A.; Weissman, I.L. Developmental cell death programs license cytotoxic cells to eliminate histocompatible partners. Proc. Natl. Acad. Sci. USA 2016, 113, 6520–6525.
- De Tomaso, A.W.; Nyholm, S.V.; Palmeri, K.J.; Ishizuka, K.J.; Ludington, W.B.; Mitchel, K.; Weissman, I.L. Isolation and characterization of a protochordate histocompatibility locus. Nature 2005, 438, 454–459.
- Uinuk-ool, T.; Mayer, W.E.; Sato, A.; Dongak, R.; Cooper, M.D.; Klein, J. Lamprey lymphocyte-like cells express homologs of genes involved in immunologically relevant activities of mammalian lymphocytes. Proc. Natl. Acad. Sci. USA 2002, 99, 14356–14361.
- Li, J.; Das, S.; Herrin, B.R.; Hirano, M.; Cooper, M.D. Definition of a third VLR gene in hagfish. Proc. Natl. Acad. Sci. USA 2013, 110, 15013–15018.
- Zou, J.; Secombes, C.J. The Function of Fish Cytokines. Biology 2016, 5, 23.
- Schwartz, J.C.; Gibson, M.S.; Heimeier, D.; Koren, S.; Phillippy, A.M.; Bickhart, D.M.; Smith, T.P.; Medrano, J.F.; Hammond, J.A. The evolution of the natural killer complex; a comparison between mammals using new high-quality genome assemblies and targeted annotation. Immunogenetics 2017, 69, 255–269.
- Shen, L.; Stuge, T.B.; Zhou, H.; Khayat, M.; Barker, K.S.; Quiniou, S.M.; Wilson, M.; Bengtén, E.; Chinchar, V.G.; Clem, L.W.; et al. Channel catfish cytotoxic cells: A mini-review. Dev. Comp. Immunol. 2002, 26, 141–149.
- Morales, H.D.; Robert, J. Characterization of primary and memory CD8 T-cell responses against ranavirus (FV3) in Xenopus laevis. J. Virol. 2007, 81, 2240–2248.
- Hirano, M.; Das, S.; Guo, P.; Cooper, M.D. The evolution of adaptive immunity in vertebrates. Adv. Immunol. 2011, 109, 125–157.
- Butenko, S.; Satyanarayanan, S.K.; Assi, S.; Schif-Zuck, S.; Barkan, D.; Sher, N.; Ariel, A. Transcriptomic Analysis of Monocyte-Derived Non-Phagocytic Macrophages Favors a Role in Limiting Tissue Repair and Fibrosis. Front. Immunol. 2020, 11, 405.
- Hanton, G.; Pastoret, P.P. The reaction of antibody-dependent cell-mediated cytotoxicity (ADCC). Ann. Vet. Res. 1984, 15, 443–454.
- Torben, W.; Ahmad, G.; Zhang, W.; Nash, S.; Le, L.; Karmakar, S.; Siddiqui, A.A. Role of antibody dependent cell mediated cytotoxicity (ADCC) in Sm-p80-mediated protection against Schistosoma mansoni. Vaccine 2012, 30, 6753–6758.
- Sun, Y.; Asmal, M.; Lane, S.; Permar, S.R.; Schmidt, S.D.; Mascola, J.R.; Letvin, N.L. Antibody-Dependent Cell-Mediated Cytotoxicity in Simian Immunodeficiency Virus-Infected Rhesus Monkeys. J. Virol. 2011, 85, 6906–6912.
- Ljunggren, H.-G.; Kärre, K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol. Today 1990, 11, 237–244.
- Raulet, D.H. Missing self recognition and self tolerance of natural killer (NK) cells. Semin. Immunol. 2006, 18, 145–150.
- Borrego, F. The First Molecular Basis of the “Missing Self” Hypothesis. J. Immunol. 2006, 177, 5759–5760.
- Khalturin, K.; Becker, M.; Rinkevich, B.; Bosch, T.C. Urochordates and the origin of natural killer cells: Identification of a CD94/NKR-P1-related receptor in blood cells of Botryllus. Proc. Natl. Acad. Sci. USA 2003, 100, 622–627.
- Kishishita, N.; Nagawa, F. Evolution of adaptive immunity: Implications of a third lymphocyte lineage in lampreys. BioEssays News Rev. Mol. Cell. Dev. Biol. 2014, 36, 244–250.
- Das, S.; Hirano, M.; Aghaallaei, N.; Bajoghli, B.; Boehm, T.; Cooper, M.D. Organization of lamprey variable lymphocyte receptor C locus and repertoire development. Proc. Natl. Acad. Sci. USA 2013, 110, 6043–6048.
- Kasahara, M. Variable Lymphocyte Receptors: A Current Overview. Results Probl. Cell Differ. 2015, 57, 175–192.
- Croft, N.P.; Smith, S.A.; Pickering, J.; Sidney, J.; Peters, B.; Faridi, P.; Witney, M.J.; Sebastian, P.; Flesch, I.E.A.; Heading, S.L.; et al. Most viral peptides displayed by class I MHC on infected cells are immunogenic. Proc. Natl. Acad. Sci. USA 2019, 116, 3112–3117.
- Marino, J.; Paster, J.; Benichou, G. Allorecognition by T Lymphocytes and Allograft Rejection. Front. Immunol. 2016, 7, 582.
- Bosselut, R. T cell antigen recognition: Evolution-driven affinities. Proc. Natl. Acad. Sci. USA 2019, 116, 21969–21971.
- Castro, R.; Bernard, D.; Lefranc, M.P.; Six, A.; Benmansour, A.; Boudinot, P. T cell diversity and TcR repertoires in teleost fish. Fish Shellfish Immunol. 2011, 31, 644–654.
- Foulkrod, A.M.; Appasamy, P.M. Expression of TCR genes in adult and larval Xenopus laevis. Dev. Comp. Immunol. 2019, 96, 78–82.




