Approximately 71 million people worldwide are infected with the hepatitis C virus (HCV). Injectable drug use represents the most common route of transmission in Europe and other developed countries. We studied the molecular characteristics of the HCV infection among mono-infected people who used drugs (PWUD) in Italy. Among 208 PWUD with anti-HCV antibodies, 101 (48.6%) were HCV RNA-positive, the majority (47%) were infected with the HCV genotype (Gt)1a, followed by Gt3a (34.9%), Gt4 (9.1%), Gt1b (4.5%), and Gt2 (4.5%). Bayesian phylogenetic analyses of clustered HCV NS5B sequences from 66 HCV-positive PWUDs with available plasma samples indicated age and neighborhood proximity as the most common characteristics between closely related HCV strains. Population dynamics, as measured by a coalescent Bayesian skyline analysis, revealed an increase in HCV Gt1a infections from the mid-1980s to mid-1990s. While HCV Gt3a infections were first detected in the 1980s, patient numbers with this genotype subtype remained relatively constant. For both Gt1a and Gt3a, Birth–Death Bayesian Skyline analyses produced higher reproduction numbers post 2014. For earlier time intervals, slow growths were observed for both Gt1a and Gt3a with reproduction numbers (Re) of approximately 1. The evolutionary rates for Gt1a and Gt3a were estimated as 2.23 × 10−4 and 3.85 × 10−4, respectively.
1. Introduction
The WHO estimated that approximately 71 million people worldwide are infected with the hepatitis C virus (HCV), with 1.75 million new infections being reported every year
[1]. This virus is a blood-borne pathogen. Before the screening for anti-HCV antibodies was introduced in 1990, blood transfusion was the main risk factor for HCV infection
[2]. In 1993, Western countries started using a nucleic acid amplification (NAT) test to detect the presence of HCV RNA. The availability of more sensitive tests led to a reduction in HCV prevalence. In Italy in 2013, the HCV RNA incidence rate in first-time blood donors was 2.5 per 100,000
[3]. Illicit drug users (IDUs) have been the predominant HCV-infected population since the early 2000s
[4]. The WHO estimated that nearly 13 million individuals worldwide are IUDs, of which 67% are living with HCV
[5]. Despite the route of transmission being similar for HCV and HIV, the transmission rates appear to vary. The HCV is estimated to be approximately ten times more transmissible than HIV, as only 30% of infected IDUs have HIV. Seroprevalence of HCV antibodies is very high among people who inject drugs (PWID); up to 90% of PWID have HCV antibodies compared with approximately 10% with HIV seroprevalence
[6][7]. Preventive strategies, such as the reduction of syringe exchange with better aseptic technique compliance, have facilitated the reduction of HCV transmission by drug injection. However, numerous studies have shown that the sharing of drug paraphernalia is responsible for the spread of HCV among both IDUs and non-IDUs
[8][9]. Specifically, the sharing of a sniffer device is a relevant factor in the spread of HCV among non-IDUs
[10][11]. Changes in drug consumption and the awareness of safer drug injection practices have reduced the incidence of HCV transmission. In France, for instance, the cumulative incidence of HCV infection in IDUs was 40–50% over the last five years with the implementation of safer drug practices; this is compared with 90% in the early 1990s
[12][13]. In Italy, in 2016, a national surveillance analysis at the Italian Centers for substance dependence indicated that approximately half a million of the population were people who use drugs (PWUD) and 30% tested positive for HCV
[14]. The HCV status of these patients remains poorly understood, especially the variability in HCV transmission dynamics among this special population. As reported for other Western countries
[15], genotypes 1a (Gt1a) and 3 (Gt3) are the most prevalent among PWID. HCV Gt4 prevalence is also increasing and drug use seems to be a key factor for the spread of this genotype
[16], combined with people emigrating from African and Asian countries. To date, no phylogenetic or phylodynamic studies have been performed that focus on the correlation of drug use with the HCV genotype and the age of the patient.
2. Current Analysis on HCV Infection
In this study, HCV infection among PWUD was evaluated around the metropolitan area of Rome. HCV RNA was detected in 48.6% of participants who tested positive for anti-HCV antibodies. This prevalence was considerably higher when compared with reports for the general population, although slightly less than reported in 2009 by Stroffolini et al. where 68.3% of HCV-positive IDUs were also viremic
[16]. We could not associate HCV RNA prevalence with specific age groups since 82% of the study participants were >45 years old. Gt1b was infrequently observed (prevalence, 4.5%) in comparison to the general population
[17], suggesting a different route of transmission. The prevalence of Gt4 (9.1%) was in line with previously described results over the past few decades in Italy and other Mediterranean basin countries
[18][19][20]. In fact, the prevalence of this genotype in Europe ranges from 8.2% to 20%
[21][22]. In France, the prevalence of Gt4 has increased from 4% in 1990 to greater than 11% over the last decade
[23][24]. In Greece, Gt4 accounts for approximately 15% of all HCV infections
[25][26]. Furthermore, our results showed a reduced prevalence of Gt4 when compared with other certain populations, such as men who have sex with men
[27].
Gt1a was the most prevalent genotype (47%) in all evaluated age categories, followed by Gt3a (34.9%). In line with our results, Stroffolini et al. reported that the most common HCV genotypes among intravenous IDUs with Gt1a and Gt3a prevalence of 47.9% and 39.7%, respectively
[16]. The marked differences between Gt1a and Gt1b strengthen the hypothesis that these two subtypes have different transmission routes. Gt1a was first introduced into Italy in the early 1970s, and coalescent Bayesian skyline analysis highlighted an exponential increase in Gt1a incidence between the 1980s and 1990s (
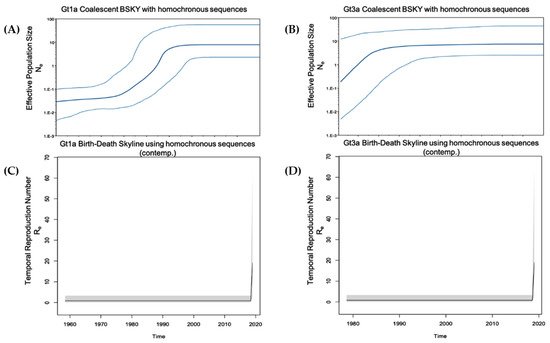
Figure 1). This finding is in accordance with the younger age of infected HCV Gt1a patients, especially when compared with Gt1b, the main genotype detected in older chronically infected HCV patients
[15]. Thus, there appears to be a dispersal origin. In the majority of cases, PWUD strains are intermixed with non-PWUD strains suggesting that Gt1a became a widely distributed subtype, affecting different patient demographics. The Gt1a sequences formed distinct clusters, distributed throughout the Bayesian phylogenetic tree without any distinct pattern. This is likely the result of a widespread and long-term distribution of Gt1a within the global population, and it is consistent with observations such as those in Greece regarding Gt1a dispersal among IDUs
[28].
Figure 1. Population growth and temporal reproduction numbers for Gt1a and Gt3a using homochronous (SerD and non-SerD) reference sequences. (A,B) The Coalescent Bayesian skyline model combined with a GTR gamma site model (4 categories) and a strict molecular clock with sequence date constraints were used to estimate effective population size. The solid middle line represents the median, while the upper and lower lines represent the 95% high posterior density interval estimate of Ne over time. (C,D) The Birth-Death Skyline (contemporary) model was used to estimate temporal reproduction numbers from homochronous sequences. Figure A/C and B/D are aligned on the same time scale (x-axis)
A higher prevalence of Gt1a was observed across all of the evaluated time intervals when compared to Gt3a (
Table 1). Gt1a prevalence was 45.2% from <1990 to 2001 and its prevalence has gradually increased through subsequent years (47.1% from 2002 to 2013 and 50% from 2014 to 2019). Gt3a accounted for 33.3% of HCV infections from <1990 to 2001, increased to 41.2% from 2002 to 2013 and decreased to 33.3% from 2014 to 2019. Gt1a prevalence was always greater than 40% for each evaluated time period, which may be related to patient age. Approximately 80% of these patients were over 45 years old and 87.9% began using drugs before 1995. Of these patients, 23 were anti-HCV antibody-positive and 12 (39.7%) were infected with Gt1a, as shown in the phylodynamic analysis, demonstrating an increase of Gt1a infection between the mid-1980s and mid-1990s. Nevertheless, since the date of the first HCV positivity is estimated, it is possible that the primary infection occurred during a time period significantly prior to the first serological test in the early years of drug use, as reported in the literature
[29]. Conversely, Gt3a prevalence ranged from 33–41% across all evaluated time intervals, in accordance with results from the coalescent Bayesian analysis illustrated in the skyline plot, which underlined a constant increase of Gt3a from 1986 onwards. The epidemiology of increased Gt3a transmission among the PWUD population is unknown. It is possible that this genotype, which is endemic in India, was introduced into Italy through PWUD who visited India in the 1970s when opium was easily accessible
[30][31]. Moreover, the increased spread of Gt3a may have been related to the migration of people, including PWUD, from Eastern Europe after the Berlin wall came down in 1989
[32]. The lower prevalence of Gt3a at all of the analysed time intervals may be due to the reduced transmission of this genotype in comparison with Gt1a or, conversely, a higher capacity to clear the primary infection
[33][34].
Table 1. Characteristics of SerD study participants with available HCV NS5B sequence and recruited between 2018 and 2019 (N.S. = not stated).
| Characteristics |
Total (n) |
HCV Genotype |
| 1a |
1b |
2 |
3a |
4 |
| Age (years), mean ± SD |
50 ± 8.9 (n = 66) |
49 ± 9.4 (n = 31) |
57 ± 3.5 (n = 3) |
61 ± 5.3 (n = 3) |
48 ± 9.1 (n = 23) |
52 ± 2.3 (n = 6) |
| ≤34 |
5 |
3 |
- |
- |
2 |
- |
| 35–44 |
7 |
4 |
- |
- |
3 |
- |
| ≥45 |
54 |
24 |
3 |
3 |
18 |
6 |
| Gender |
(n = 66) |
(n = 31) |
(n = 3) |
(n = 3) |
(n = 23) |
(n = 6) |
| Male |
54 |
26 |
3 |
3 |
18 |
4 |
| Female |
12 |
5 |
- |
- |
5 |
2 |
| Estimated year of HCV infection |
(n = 66) |
(n = 31) |
(n = 3) |
(n = 3) |
(n = 23) |
(n = 6) |
| <1990 |
7 |
5 |
- |
- |
1 |
1 |
| 1990–1995 |
22 |
8 |
1 |
1 |
9 |
3 |
| 1996–2001 |
13 |
6 |
1 |
1 |
4 |
1 |
| 2002–2007 |
7 |
1 |
1 |
- |
4 |
1 |
| 2008–2013 |
10 |
7 |
- |
- |
3 |
- |
| 2014–2019 |
6 |
3 |
- |
1 |
2 |
- |
| NS |
1 |
1 |
- |
- |
- |
- |
| Assumption for infection |
(n = 66) |
(n = 31) |
(n = 3) |
(n = 3) |
(n = 23) |
(n = 6) |
| Inhalation |
4 |
2 |
- |
- |
1 |
1 |
| Intravenous |
5 |
4 |
- |
- |
1 |
- |
| Inhal and Intrav |
55 |
23 |
3 |
3 |
21 |
5 |
| NS |
2 |
2 |
- |
|
- |
- |
| Most commonly injected drug at recruitment |
(n = 66) |
(n = 31) |
(n = 3) |
(n = 3) |
(n = 23) |
(n = 6) |
| Heroin |
20 |
9 |
- |
2 |
8 |
1 |
| Cocaine |
1 |
1 |
- |
- |
- |
- |
| Heroin and Cocaine |
45 |
21 |
3 |
1 |
15 |
5 |
By applying ‘Birth and Death Skyline contemporary (BDSKYcontemp)’ and ‘Birth and Death Skyline serial (BDSKYserial)’ analyses for homochronous (SerD and non-SerD cohorts) and heterochronous (includes non-Italian references) sequences, respectively, the effective temporal Re could be estimated. When using homochronous sequences and applying a BDSKYcontemp model, there was no algorithm calibration based on sample dates, however, the analysis focused solely on query sequences. By including heterochronous sequences and applying a BDSKYserial model, there was greater confidence in the generated numbers, with the caveat that the results may have partially reflected population dynamics from the reference sequences. Both analyses indicated an overall constant population (Re ≈ 1), culminating in a very recent sharp increase. While this might be qualitatively true, Re numbers obtained using the BDSKYcontemp model were probably inflated, while Re calculated using the BDSKYserial model were conservative. Qualitatively, both analyses showed an increase in HCV infections over the past five years.
The phylodynamic analysis was valuable in the identification of different clusters and transmission routes among PWUD, as with the participants infected with Gt2b
[35][36]. These findings could not be extracted from traditional epidemiological analyses. The anti-HCV antibody-positivity for these Gt2b participants was established during different time intervals (between 1995 and 2019). Nevertheless, as stated in the questionnaire, the participants were friends who exchanged syringes while injecting heroin or cocaine. A limitation of the phylogenetic and phylodynamic analyses was the inability to identify whether a particular strain had a higher risk of transmission. Moreover, it is unknown whether other HCV-positive individuals registered at each SerD, but were not a part of our study, were infected with similar HCV strains and either recovered or progressed to chronic infection. Access to this type of information can only be obtained in a prospective study. Interestingly, none of our study sequences (beginning with either INMI, Pt, or Q) clustered with sequences from other countries. A possible explanation is the limited sample size.