
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mark Mellett | + 3446 word(s) | 3446 | 2021-05-08 08:26:24 | | | |
| 2 | Dean Liu | -10 word(s) | 3436 | 2021-06-29 03:28:24 | | |
Video Upload Options
The nucleotide-binding domain and leucine-rich-repeat-containing family (NLRs) (sometimes called the NOD-like receptors, though the family contains few bona fide receptors) are a superfamily of multidomain-containing proteins that detect cellular stress and microbial infection.
1. Introduction
Innate immunity relies on the recognition of evolutionarily conserved microbe-specific molecules, termed pathogen-associated molecular patterns (PAMPs). Germline encoded pattern recognition receptors (PRRs) expressed on the cell surface, endosomes or in the cytosol detect and respond to these PAMPs. Although, the domains of these PRRs are highly conserved, extensive species-specific expansions and domain shuffling result in an advantage to an organism living in pathogen-rich environments. The PRRs expressed by mammalian cells are Toll-like receptors (TLRs), the NOD-like receptors (NLRs), AIM2-like receptors (ALRs), RIG-like receptors (RLRs) and C-type lectin receptors (CLRs), with each family member recognizing specific molecular signatures [1]. Two of these families of PRRs are conserved from early invertebrates to mammals: the transmembrane TLRs and the intracellular NLRs [2][3].
Our skin acts as a sentinel organ, determining when and how to respond to a broad range of environmental insults during both homeostatic and pathologic situations. The skin forms a physical barrier through the cornified envelope of stratum corneum and via tight-junctions in lower layers, a chemical barrier by maintaining an acidic pH and antimicrobial peptide expression and finally, there is the immunologic barrier formed by keratinocytes and infiltrating immune cells of both the innate and adaptive immune systems [4]. These layers of barriers interact with each other to protect the organism from harmful stimuli. Keratinocytes are the main cell type of the epidermis and as immunocompetent cells are implicated in the protection against harmful threats, by the expression of a wide range of PRRs, including TLRs and NLRs [5][6][7]. The activation of PRRs induces keratinocytes to express antimicrobial peptides and immune mediators, which promote the recruitment of professional immune cells [4]. Murine and human TLRs in skin biology have been discussed elsewhere [5][6], here we will focus on the NLR family and discuss its role in the immune defence in the skin.
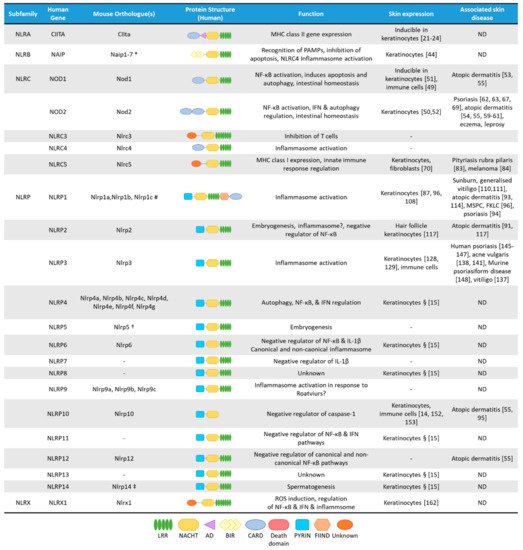
NLRs are cytosolic receptors widely identified in non-vertebrates and vertebrates, and have functional analogues, the R-proteins, in plants [8]. The number of NLR genes differ from species to species: humans express 23 NLR family members, while mice have at least 34 NLR paralogues (Figure 1) [1][8]. NLRs are multi-domain containing proteins, comprising of a C-terminal domain with a series of leucine rich repeats (LRRs), a central nucleotide-binding NACHT domain, and an N-terminal effector domain [9]. The N-terminal domain is variable, and NLRs are divided into five subfamilies based on their distinctive N-terminal domain: NLRAs that have an acidic activation domain, NLRBs that possess a baculovirus inhibitor of apoptosis repeat (BIR)-like domain, NLRCs that feature a caspase activation and recruitment domain (CARD) or a Death domain (DD), and the NLRP subfamily that contain a PYRIN domain [9]. The NLRX subfamily contains one member, and its nomenclature derives from an uncharacterized N-terminal domain that lacks homology with other NLR effector domains.
In the cytosol, NLRs remain in an auto-inhibitory state. The LRR domains are thought to be responsible for ligand binding but this has not been experimentally shown to date for most NLRs and this dearth of evidence has led to the belief that mammalian LRRs might not have necessarily retained this function [10][11]. The LRR region also maintains the NLR in an auto-inhibitory state, as demonstrated by the crystal structure of NLRC4, where the LRR obstructs the NACHT domain [12]. The NACHT domain possesses dNTPase activity, which governs the ATP-dependent oligomerisation. Although, the NACHT domain controls oligomerisation, additionally ligand-binding can occur in this region. Upon activation, the N-terminal domain activates distinctive downstream signalling cascades resulting in an inflammatory response. This innate immune response also serves to influence the adaptive arm of the immune system [1]. Despite subfamilies sharing the same domain, individual members can elicit different downstream effects. For example, the NLRC family that contains a CARD domain, induces inflammasome activation, regulates nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB) or type I interferon (IFN) signalling pathways or engages in transcriptional regulation [13].
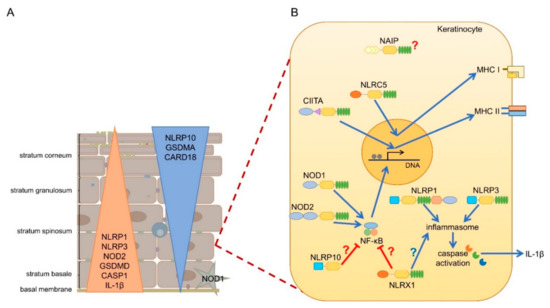
mRNA expression of most NLRs are found in the skin, but since NLR activation is a very complex process, and some NLRs are characterized by unique cell-type specific features, without their functional evaluation in keratinocytes, their functions cannot be clearly addressed. Cornification of keratinocytes also affects the expression of NLRs and their interacting partners (Figure 2A) [14]. Here, we will summarize the current knowledge on epidermal NLR expression and functions and their potential contribution to skin disease.
2. The NLRA Subfamily
3. The NLRB Subfamily
4. The NLRC Subfamily
4.1. NOD1 and NOD2
4.2. NLRC5
4.3. Skin Diseases Associated with NLRC5 Functions
5. The NLRP Subfamily
5.1. NLRP1
5.2. NLRP2
5.3. NLRP3
5.4. NLRP10
6. The NLRX Subfamily
The sole member of the NLRX family, NLRX1 contains a dissimilar and uncharacterised N-terminal effector domain compared to other NLRs. It also has an unusual C-terminus, which contains seven LRRs and a three-helix bundle [121]. Within the N-terminus NLRX1 contains a mitochondria-targeting sequence [122][123][124] and is involved in mitochondrial reactive oxygen species (ROS) formation [124]. Additionally, NLRX1 attenuates NF-κB and inflammasome signalling [124][125]. The regulatory effects of NLRX1 are highly cell type specific, which might be determined by the unique functional activity or metabolic profile of the given cell type [126]. NLRX1 is ubiquiteously expressed, including in keratinocytes, but its function in the skin is currently unknown.
References
- Chandler, C.E.; Harberts, E.M.; Ernst, R.K. Pathogen Sensing: Toll-Like Receptors and NODs (Innate Immunity). In Encyclopedia of Microbiology (Fourth Edition); Elsevier: Amsterdam, The Netherlands, 2019; pp. 443–456.
- Zhang, Q.; Zmasek, C.M.; Godzik, A. Domain architecture evolution of pattern-recognition receptors. Immunogenetics 2010, 62, 263–272.
- Jann, O.C.; King, A.; Lopez Corrales, N.; Anderson, S.I.; Jensen, K.; Ait-ali, T.; Tang, H.; Wu, C.; Cockett, N.E.; Archibald, A.L.; et al. Comparative genomics of Toll-like receptor signalling in five species. BMC Genom. 2009, 10, 1–15.
- Kuo, I.-H.; Yoshida, T.; De Benedetto, A.; Beck, L.A. The cutaneous innate immune response in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2013, 131, 266–278.
- Sun, L.; Liu, W.; Zhang, L.J. The Role of Toll-Like Receptors in Skin Host Defense, Psoriasis, and Atopic Dermatitis. J. Immunol. Res. 2019, 1824624.
- Miller, L.S. Toll-like receptors in skin. Adv. Dermatol. 2008, 24, 71–87.
- Tang, L.; Zhou, F. Inflammasomes in Common Immune-Related Skin Diseases. Front. Immunol. 2020, 11, 1–15.
- Shaw, M.H.; Reimer, T.; Kim, Y.-G.; Nuñez, G. NOD-like receptors (NLRs): Bona fide intracellular microbial sensors. Curr. Opin. Immunol. 2008, 20, 377–382.
- Ting, J.P.Y.; Lovering, R.C.; Alnemri, E.S.P.D.; Bertin, J.; Boss, J.M.; Davis, B.; Flavell, R.A.; Girardin, S.E.; Godzik, A.; Harton, J.A.; et al. The NLR gene family: An official nomenclature. Immunity 2008, 28, 285–287.
- Bentham, A.; Burdett, H.; Anderson, P.A.; Williams, S.J.; Kobe, B. Animal NLRs provide structural insights into plant NLR function. Ann. Bot. 2016, 119, 698–702.
- Tenthorey, J.L.; Kofoed, E.M.; Daugherty, M.D.; Malik, H.S.; Vance, R.E. Molecular Basis for Specific Recognition of Bacterial Ligands by NAIP/NLRC4 Inflammasomes. Mol. Cell 2014, 54, 17–29.
- Hu, Z.; Yan, C.; Liu, P.; Huang, Z.; Ma, R.; Zhang, C.; Wang, R.; Zhang, Y.; Martinon, F.; Miao, D.; et al. Crystal Structure of NLRC4 Reveals Its Autoinhibition Mechanism. Science 2013, 341, 172–175.
- Meunier, E.; Broz, P. Evolutionary Convergence and Divergence in NLR Function and Structure. Trends Immunol. 2017, 38, 744–757.
- Lachner, J.; Mlitz, V.; Tschachler, E.; Eckhart, L. Epidermal cornification is preceded by the expression of a keratinocyte-specific set of pyroptosis-related genes. Sci. Rep. 2017, 7, 17446.
- Watanabe, H.; Gaide, O.; Pétrilli, V.; Martinon, F.; Contassot, E.; Roques, S.; Kummer, J.A.; Tschopp, J.; French, L.E. Activation of the IL-1beta-processing inflammasome is involved in contact hypersensitivity. J. Investig. Dermatol. 2007, 127, 1956–1963.
- León Machado, J.A.; Steimle, V. The MHC class II transactivator CIITA: Not (quite) the odd-one-out anymore among NLR proteins. Int. J. Mol. Sci. 2021, 22, 1074.
- Steimle, V.; Otten, L.A.; Zufferey, M.; Mach, B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome). Cell 1993, 75, 135–146.
- Chang, C.-H.; Guerder, S.; Hong, S.-C.; van Ewijk, W.; Flavell, R.A. Mice Lacking the MHC Class II Transactivator (CIITA) Show Tissue-Specific Impairment of MHC Class II Expression. Immunity 1996, 4, 167–178.
- Reith, W.; LeibundGut-Landmann, S.; Waldburger, J.-M. Regulation of MHC class II gene expression by the class II transactivator. Nat. Rev. Immunol. 2005, 5, 793–806.
- Choi, N.M.; Majumder, P.; Boss, J.M. Regulation of major histocompatibility complex class II genes. Curr. Opin. Immunol. 2011, 23, 81–87.
- Albanesi Cavani, C.A.; Girolomoni, G. Interferon-γ-Stimulated Human Keratinocytes Express the Genes Necessary for the Production of Peptide-Loaded MHC Class II Molecules. J. Investig. Dermatol. 1998, 110, 138–142.
- Takagi, A.; Nishiyama, C.; Kanada, S.; Niwa, Y.; Fukuyama, K.; Ikeda, S.; Okumura, K.; Ogawa, H. Prolonged MHC class II expression and CIITA transcription in human keratinocytes. Biochem. Biophys. Res. Commun. 2006, 347, 388–393.
- Tamoutounour, S.; Han, S.-J.; Deckers, J.; Constantinides, M.G.; Hurabielle, C.; Harrison, O.J.; Bouladoux, N.; Linehan, J.L.; Link, V.M.; Vujkovic-Cvijin, I.; et al. Keratinocyte-intrinsic MHCII expression controls microbiota-induced Th1 cell responses. Proc. Natl. Acad. Sci. USA 2019, 116, 23643–23652.
- Czernielewski, J.M.; Bagot, M. Class II MHC antigen expression by human keratinocytes results from lympho-epidermal interactions and gamma-interferon production. Clin. Exp. Immunol. 1986, 66, 295–302.
- Aubock, J.; Romani, N.; Grubauer, G.; Fritsch, P. HLA-DR expression on keratinocytes is a common feature of diseased skin. Br. J. Dermatol. 1986, 114, 465–472.
- Krueger, J.G.; Krane, J.F.; Carter, D.M.; Gottlieb, A.B. Role of Growth Factors, Cytokines, and Their Receptors in the Pathogenesis of Psoriasis. J. Investig. Dermatol. 1990, 94, s135–s140.
- Wittmann, M.; Purwar, R.; Hartmann, C.; Gutzmer, R.; Werfel, T. Human Keratinocytes Respond to Interleukin-18: Implication for the Course of Chronic Inflammatory Skin Diseases. J. Investig. Dermatol. 2005, 124, 1225–1233.
- Mutis, T.; Bueger, M.; Bakker, A.; Ottenhoff, T.H.M. HLA Class II+ Human Keratinocytes present Mycobacterium leprae Antigens to CD4+ Thl-Like Cells. Scand. J. Immunol. 1993, 37, 43–51.
- Nickoloff, B.J.; Basham, T.Y.; Merigan, T.C.; Torseth, J.W.; Morhenn, V.B. Human Keratinocyte-Lymphocyte Reactions In Vitro. J. Investig. Dermatol. 1986, 87, 11–18.
- Gaspari, A.A.; Katz, S.I. Induction and functional characterization of class II MHC (Ia) antigens on murine keratinocytes. J. Immunol. 1988, 140, 2956–2963.
- Niederwieser, D.; Auböck, J.; Troppmair, J.; Herold, M.; Schuler, G.; Boeck, G.; Lotz, J.; Fritsch, P.; Huber, C. IFN-mediated induction of MHC antigen expression on human keratinocytes and its influence on in vitro alloimmune responses. J. Immunol. 1988, 140, 2556–2564.
- Ortiz-Sanchez, E.; Chávez-Olmos, P.; Pina-Sanchez, P.; Salcedo, M.; Garrigo, E. Expression of the costimulatory molecule CD86, but not CD80, in keratinocytes of normal cervical epithelium and human papillomavirus-16 positive low squamous intraepithelial lesions. Int. J. Gynecol. Cancer 2007, 17, 571–580.
- Nickoloff, B.J.; Mitra, R.S.; Lee, K.; Turka, L.A.; Green, J.; Thompson, C.; Shimizu, Y. Discordant expression of CD28 ligands, BB-1, and B7 on keratinocytes in vitro and psoriatic cells in vivo. Am. J. Pathol. 1993, 142, 1029–1040.
- Gaspari, A.A.; Ferbel, B.; Chen, Z.; Razvi, F.; Polakowska, R. Accessory and Alloantigen-Presenting Cell Functions of A431 Keratinocytes That Stably Express the B7 Antigen. Cell. Immunol. 1993, 149, 291–302.
- Nickoloff, B.J.; Turka, L.A. Immunological functions of non-professional antigen-presenting cells: New insights from studies of T-cell interactions with keratinocytes. Immunol. Today 1994, 15, 464–469.
- Kofoed, E.M.; Vance, R.E. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 2011, 477, 592–595.
- Maier, J.K.X.; Lahoua, Z.; Gendron, N.H.; Fetni, R.; Johnston, A.; Davoodi, J.; Rasper, D.; Roy, S.; Slack, R.S.; Nicholson, D.W.; et al. The Neuronal Apoptosis Inhibitory Protein Is a Direct Inhibitor of Caspases 3 and 7. J. Neurosci. 2002, 22, 2035–2043.
- Davoodi, J.; Ghahremani, M.-H.; Es-haghi, A.; Mohammad-gholi, A.; MacKenzie, A. Neuronal apoptosis inhibitory protein, NAIP, is an inhibitor of procaspase-9. Int. J. Biochem. Cell Biol. 2010, 42, 958–964.
- Sanna, M.G.; da Correia, J.S.; Ducrey, O.; Lee, J.; Nomoto, K.; Schrantz, N.; Deveraux, Q.L.; Ulevitch, R.J. IAP Suppression of Apoptosis Involves Distinct Mechanisms: The TAK1/JNK1 Signaling Cascade and Caspase Inhibition. Mol. Cell. Biol. 2002, 22, 1754–1766.
- Vance, R.E. The NAIP/NLRC4 inflammasomes. Curr. Opin. Immunol. 2015, 32, 84–89.
- Yang, J.; Zhao, Y.; Shi, J.; Shao, F. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl. Acad. Sci. USA 2013, 110, 14408–14413.
- Mariathasan, S.; Newton, K.; Monack, D.M.; Vucic, D.; French, D.M.; Lee, W.P.; Roose-Girma, M.; Erickson, S.; Dixit, V.M. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 2004, 430, 213–218.
- Zamboni, D.S.; Kobayashi, K.S.; Kohlsdorf, T.; Ogura, Y.; Long, E.M.; Vance, R.E.; Kuida, K.; Mariathasan, S.; Dixit, V.M.; Flavell, R.A.; et al. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat. Immunol. 2006, 7, 318–325.
- Human Protein Atlas NAIP. Available online: (accessed on 19 February 2021).
- Velloso, F.J.; Trombetta-Lima, M.; Anschau, V.; Sogayar, M.C.; Correa, R.G. NOD-like receptors: Major players (and targets) in the interface between innate immunity and cancer. Biosci. Rep. 2019, 39.
- Human Protein Atlas. Available online: (accessed on 31 January 2021).
- Correa, R.G.; Milutinovic, S.; Reed, J.C. Roles of NOD1 (NLRC1) and NOD2 (NLRC2) in innate immunity and inflammatory diseases. Biosci. Rep. 2012, 32, 597–608.
- Travassos, L.H.; Carneiro, L.A.M.; Girardin, S.; Philpott, D.J. Nod proteins link bacterial sensing and autophagy. Autophagy 2010, 6, 409–411.
- Human Protein Atlas NOD1. Available online: (accessed on 19 February 2021).
- Human Protein Atlas NOD2. Available online: (accessed on 19 February 2021).
- Harder, J.; Núñez, G. Functional Expression of the Intracellular Pattern Recognition Receptor NOD1 in Human Keratinocytes. J. Investig. Dermatol. 2009, 129, 1299–1302.
- Voss, E.; Wehkamp, J.; Wehkamp, K.; Stange, E.F.; Schröder, J.M.; Harder, J. NOD2/CARD15 mediates induction of the antimicrobial peptide human beta-defensin-2. J. Biol. Chem. 2006, 281, 2005–2011.
- Human Protein Atlas NLRC5. Available online: (accessed on 19 February 2021).
- Mótyán, J.; Bagossi, P.; Benkő, S.; Tőzsér, J. A molecular model of the full-length human NOD-like receptor family CARD domain containing 5 (NLRC5) protein. BMC Bioinform. 2013, 14, 275.
- Downs, I.; Vijayan, S.; Sidiq, T.; Kobayashi, K.S. CITA/NLRC5: A critical transcriptional regulator of MHC class I gene expression. BioFactors 2016, 42, 349–357.
- Neerincx, A.; Castro, W.; Guarda, G.; Kufer, T.A. NLRC5, at the Heart of Antigen Presentation. Front. Immunol. 2013, 4.
- Kobayashi, K.S.; van den Elsen, P.J. NLRC5: A key regulator of MHC class I-dependent immune responses. Nat. Rev. Immunol. 2012, 12, 813–820.
- Neerincx, A.; Jakobshagen, K.; Utermöhlen, O.; Büning, H.; Steimle, V.; Kufer, T.A. The N-Terminal Domain of NLRC5 Confers Transcriptional Activity for MHC Class I and II Gene Expression. J. Immunol. 2014, 193, 3090–3100.
- Benkő, S.; Kovács, E.G.; Hezel, F.; Kufer, T.A. NLRC5 Functions beyond MHC I Regulation—What Do We Know So Far? Front. Immunol. 2017, 8.
- Cui, J.; Zhu, L.; Xia, X.; Wang, H.Y.; Legras, X.; Hong, J.; Ji, J.; Shen, P.; Zheng, S.; Chen, Z.J.; et al. NLRC5 Negatively Regulates the NF-κB and Type I Interferon Signaling Pathways. Cell 2010, 141, 483–496.
- Benko, S.; Magalhaes, J.G.; Philpott, D.J.; Girardin, S.E. NLRC5 Limits the Activation of Inflammatory Pathways. J. Immunol. 2010, 185, 1681–1691.
- Kuenzel, S.; Till, A.; Winkler, M.; Häsler, R.; Lipinski, S.; Jung, S.; Grötzinger, J.; Fickenscher, H.; Schreiber, S.; Rosenstiel, P. The Nucleotide-Binding Oligomerization Domain-Like Receptor NLRC5 Is Involved in IFN-Dependent Antiviral Immune Responses. J. Immunol. 2010, 184, 1990–2000.
- Neerincx, A.; Lautz, K.; Menning, M.; Kremmer, E.; Zigrino, P.; Hösel, M.; Büning, H.; Schwarzenbacher, R.; Kufer, T.A. A Role for the Human Nucleotide-binding Domain, Leucine-rich Repeat-containing Family Member NLRC5 in Antiviral Responses*. J. Biol. Chem. 2010, 285, 26223–26232.
- Ranjan, P.; Singh, N.; Kumar, A.; Neerincx, A.; Kremmer, E.; Cao, W.; Davis, W.G.; Katz, J.M.; Gangappa, S.; Lin, R.; et al. NLRC5 interacts with RIG-I to induce a robust antiviral response against influenza virus infection. Eur. J. Immunol. 2015, 45, 758–772.
- Davis, B.K.; Roberts, R.A.; Huang, M.T.; Willingham, S.B.; Conti, B.J.; Brickey, W.J.; Barker, B.R.; Kwan, M.; Taxman, D.J.; Accavitti-Loper, M.-A.; et al. Cutting Edge: NLRC5-Dependent Activation of the Inflammasome. J. Immunol. 2011, 186, 1333–1337.
- Spoerri, I.; Herms, S.; Eytan, O.; Sair, O.; Heinimann, K.; Sprecher, E.; Itin, P.H.; Burger, B. Immune-regulatory genes as possible modifiers of familial pityriasis rubra pilaris—Lessons from a family with PRP and psoriasis. J. Eur. Acad. Dermatol. Venereol. 2018.
- Kim, H.; Kim, H.; Feng, Y.; Li, Y.; Tamiya, H.; Tocci, S.; Ronai, Z.A. PRMT5 control of cGAS/STING and NLRC5 pathways defines melanoma response to antitumor immunity. Sci. Transl. Med. 2020, 12, eaaz5683.
- Ma, H.-L.; Zhao, X.-F.; Chen, G.-Z.; Fang, R.-H.; Zhang, F.-R. Silencing NLRC5 inhibits extracellular matrix expression in keloid fibroblasts via inhibition of transforming growth factor-β1/Smad signaling pathway. Biomed. Pharmacother. 2016, 83, 1016–1021.
- Corridoni, D.; Arseneau, K.O.; Cifone, M.G.; Cominelli, F. The Dual Role of Nod-Like Receptors in Mucosal Innate Immunity and Chronic Intestinal Inflammation. Front. Immunol. 2014, 5, 317.
- Sand, J.; Haertel, E.; Biedermann, T.; Contassot, E.; Reichmann, E.; French, L.E.; Werner, S.; Beer, H.-D. Expression of inflammasome proteins and inflammasome activation occurs in human, but not in murine keratinocytes. Cell Death Dis. 2018, 9, 24.
- Niebuhr, M.; Baumert, K.; Heratizadeh, A.; Satzger, I.; Werfel, T. Impaired NLRP3 inflammasome expression and function in atopic dermatitis due to Th2 milieu. Allergy 2014.
- Schroder, K.; Tschopp, J. The inflammasomes. Cell 2010, 140, 821–832.
- Feldmeyer, L.; Keller, M.; Niklaus, G.; Hohl, D.; Werner, S.; Beer, H.-D. The inflammasome mediates UVB-induced activation and secretion of interleukin-1beta by keratinocytes. Curr. Biol. 2007, 17, 1140–1145.
- Thürmann, L.; Grützmann, K.; Klös, M.; Bieg, M.; Winter, M.; Polte, T.; Bauer, T.; Schick, M.; Bewerunge-Hudler, M.; Röder, S.; et al. Early-onset childhood atopic dermatitis is related to NLRP2 repression. J. Allergy Clin. Immunol. 2018, 141, 1482–1485.e16.
- Levandowski, C.B.; Mailloux, C.M.; Ferrara, T.M.; Gowan, K.; Ben, S.; Jin, Y.; McFann, K.K.; Holland, P.J.; Fain, P.R.; Dinarello, C.A.; et al. NLRP1 haplotypes associated with vitiligo and autoimmunity increase interleukin-1 processing via the NLRP1 inflammasome. Proc. Natl. Acad. Sci. USA 2013, 110, 2952–2956.
- Bivik, C.; Verma, D.; Winge, M.C.; Lieden, A.; Bradley, M.; Rosdahl, I.; Söderkvist, P. Genetic Variation in the Inflammasome and Atopic Dermatitis Susceptibility. J. Investig. Dermatol. 2013, 133, 2486–2489.
- Ekman, A.-K.; Verma, D.; Fredrikson, M.; Bivik, C.; Enerbäck, C. Genetic variations of NLRP1: Susceptibility in psoriasis. Br. J. Dermatol. 2014, 171, 1517–1520.
- Hirota, T.; Takahashi, A.; Kubo, M.; Tsunoda, T.; Tomita, K.; Sakashita, M.; Yamada, T.; Fujieda, S.; Tanaka, S.; Doi, S.; et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat. Genet. 2012, 44, 1222–1226.
- Zhong, F.L.; Mamaï, O.; Sborgi, L.; Boussofara, L.; Hopkins, R.; Robinson, K.; Szeverényi, I.; Takeichi, T.; Balaji, R.; Lau, A.; et al. Germline NLRP1 Mutations Cause Skin Inflammatory and Cancer Susceptibility Syndromes via Inflammasome Activation. Cell 2016, 167, 187–202.e17.
- Martinon, F.; Burns, K.; Tschopp, J. The Inflammasome. Mol. Cell 2002, 10, 417–426.
- Zhong, Y.; Kinio, A.; Saleh, M. Functions of NOD-Like Receptors in Human Diseases. Front. Immunol. 2013, 4, 1–18.
- Faustin, B.; Lartigue, L.; Bruey, J.-M.; Luciano, F.; Sergienko, E.; Bailly-Maitre, B.; Volkmann, N.; Hanein, D.; Rouiller, I.; Reed, J.C. Reconstituted NALP1 Inflammasome Reveals Two-Step Mechanism of Caspase-1 Activation. Mol. Cell 2007, 25, 713–724.
- Boyden, E.D.; Dietrich, W.F. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat. Genet. 2006, 38, 240–244.
- Mitchell, P.S.; Sandstrom, A.; Vance, R.E. The NLRP1 inflammasome: New mechanistic insights and unresolved mysteries. Curr. Opin. Immunol. 2019, 60, 37–45.
- Sandstrom, A.; Mitchell, P.S.; Goers, L.; Mu, E.W.; Lesser, C.F.; Vance, R.E. Functional degradation: A mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 2019, 364, eaau1330.
- Chavarría-Smith, J.; Vance, R.E. Direct Proteolytic Cleavage of NLRP1B Is Necessary and Sufficient for Inflammasome Activation by Anthrax Lethal Factor. PLoS Pathog. 2013, 9, e1003452.
- Hellmich, K.A.; Levinsohn, J.L.; Fattah, R.; Newman, Z.L.; Maier, N.; Sastalla, I.; Liu, S.; Leppla, S.H.; Moayeri, M. Anthrax Lethal Factor Cleaves Mouse Nlrp1b in Both Toxin-Sensitive and Toxin-Resistant Macrophages. PLoS ONE 2012, 7, e49741.
- Levinsohn, J.L.; Newman, Z.L.; Hellmich, K.A.; Fattah, R.; Getz, M.A.; Liu, S.; Sastalla, I.; Leppla, S.H.; Moayeri, M. Anthrax Lethal Factor Cleavage of Nlrp1 Is Required for Activation of the Inflammasome. PLoS Pathog. 2012, 8, e1002638.
- Ewald, S.E.; Chavarria-Smith, J.; Boothroyd, J.C. NLRP1 Is an Inflammasome Sensor for Toxoplasma gondii. Infect. Immun. 2014, 82, 460–468.
- Bauernfried, S.; Scherr, M.J.; Pichlmair, A.; Duderstadt, K.E.; Hornung, V. Human NLRP1 is a sensor for double-stranded RNA. Science 2021, 371, eabd0811.
- Fenini, G.; Karakaya, T.; Hennig, P.; Di Filippo, M.; Beer, H.D. The NLRP1 inflammasome in human skin and beyond. Int. J. Mol. Sci. 2020, 21, 4788.
- Bachovchin, D.A. NLRP1: A jack of all trades, or a master of one? Mol. Cell 2021, 81, 423–425.
- Bruey, J.M.; Bruey-Sedano, N.; Newman, R.; Chandler, S.; Stehlik, C.; Reed, J.C. PAN1/NALP2/PYPAF2, an Inducible Inflammatory Mediator That Regulates NF-κB and Caspase-1 Activation in Macrophages. J. Biol. Chem. 2004, 279, 51897–51907.
- Ji, S.; Shin, J.E.; Kim, Y.S.; Oh, J.-E.; Min, B.-M.; Choi, Y. Toll-Like Receptor 2 and NALP2 Mediate Induction of Human Beta-Defensins by Fusobacterium nucleatum in Gingival Epithelial Cells. Infect. Immun. 2009, 77, 1044–1052.
- Netea, M.G.; Nold-Petry, C.A.; Nold, M.F.; Joosten, L.A.B.; Opitz, B.; van der Meer, J.H.M.; van de Veerdonk, F.L.; Ferwerda, G.; Heinhuis, B.; Devesa, I.; et al. a Differential requirement for the activation of the inflammasome for processing and release of IL-1beta in monocytes and macrophages. Blood 2009, 113, 2324–2335.
- Pétrilli, V.; Papin, S.; Dostert, C.; Mayor, A.; Martinon, F.; Tschopp, J. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 2007, 14, 1583–1589.
- Martinon, F.; Pétrilli, V.; Mayor, A.; Tardivel, A.; Tschopp, J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 2006, 440, 237–241.
- Martinon, F.; Agostini, L.; Meylan, E.; Tschopp, J. Identification of Bacterial Muramyl Dipeptide as Activator of the NALP3/Cryopyrin Inflammasome. Curr. Biol. 2004, 14, 1929–1934.
- Kanneganti, T.-D.; Body-Malapel, M.; Amer, A.; Park, J.-H.; Whitfield, J.; Franchi, L.; Taraporewala, Z.F.; Miller, D.; Patton, J.T.; Inohara, N.; et al. Critical Role for Cryopyrin/Nalp3 in Activation of Caspase-1 in Response to Viral Infection and Double-stranded RNA. J. Biol. Chem. 2006, 281, 36560–36568.
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420.
- He, W.; Wan, H.; Hu, L.; Chen, P.; Wang, X.; Huang, Z.; Yang, Z.-H.; Zhong, C.-Q.; Han, J. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 2015, 25, 1285–1298.
- Rühl, S.; Broz, P. Caspase-11 activates a canonical NLRP3 inflammasome by promoting K + efflux. Eur. J. Immunol. 2015, 45, 2927–2936.
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660–665.
- Grossi, S.; Fenini, G.; Kockmann, T.; Hennig, P.; Di Filippo, M.; Beer, H.-D. Inactivation of the Cytoprotective Major Vault Protein by Caspase-1 and -9 in Epithelial Cells during Apoptosis. J. Investig. Dermatol. 2020, 140, 1335–1345.e10.
- Zhuang, T.; Li, S.; Yi, X.; Guo, S.; Wang, Y.; Chen, J.; Liu, L.; Jian, Z.; Gao, T.; Kang, P.; et al. Tranilast Directly Targets NLRP3 to Protect Melanocytes from Keratinocyte-Derived IL-1β Under Oxidative Stress. Front. Cell Dev. Biol. 2020, 8, 588.
- Dai, X.; Tohyama, M.; Murakami, M.; Sayama, K. Epidermal keratinocytes sense dsRNA via the NLRP3 inflammasome, mediating interleukin (IL)-1β and IL-18 release. Exp. Dermatol. 2017, 26, 904–911.
- Zhang, C.; Xiao, C.; Dang, E.; Cao, J.; Zhu, Z.; Fu, M.; Yao, X.; Liu, Y.; Jin, B.; Wang, G.; et al. CD100–Plexin-B2 Promotes the Inflammation in Psoriasis by Activating NF-κB and the Inflammasome in Keratinocytes. J. Investig. Dermatol. 2018, 138, 375–383.
- Hasegawa, T.; Nakashima, M.; Suzuki, Y. Nuclear DNA damage-triggered NLRP3 inflammasome activation promotes UVB-induced inflammatory responses in human keratinocytes. Biochem. Biophys. Res. Commun. 2016, 477, 329–335.
- Ito, H.; Kanbe, A.; Sakai, H.; Seishima, M. Activation of NLRP3 signalling accelerates skin wound healing. Exp. Dermatol. 2018, 27, 80–86.
- Weinheimer-Haus, E.M.; Mirza, R.E.; Koh, T.J. Nod-Like Receptor Protein-3 Inflammasome Plays an Important Role during Early Stages of Wound Healing. PLoS ONE 2015, 10, e0119106.
- Wang, T.; Zhao, J.; Zhang, J.; Mei, J.; Shao, M.; Pan, Y.; Yang, W.; Jiang, Y.; Liu, F.; Jia, W. Heparan sulfate inhibits inflammation and improves wound healing by downregulating the NLR family pyrin domain containing 3 (NLRP3) inflammasome in diabetic rats. J. Diabetes 2018, 10, 556–563.
- Qing, L.; Fu, J.; Wu, P.; Zhou, Z.; Yu, F.; Tang, J. Metformin induces the m2 macrophage polarization to accelerate the wound healing via regulating ampk/mtor/nlrp3 inflammasome singling pathway. Am. J. Transl. Res. 2019, 11, 655–668.
- Lautz, K.; Damm, A.; Menning, M.; Wenger, J.; Adam, A.C.; Zigrino, P.; Kremmer, E.; Kufer, T.A. NLRP10 enhances shigella-induced pro-inflammatory responses. Cell. Microbiol. 2012, 14, 1568–1583.
- Imamura, R.; Wang, Y.; Kinoshita, T.; Suzuki, M.; Noda, T.; Sagara, J.; Taniguchi, S.; Okamoto, H.; Suda, T. Anti-Inflammatory Activity of PYNOD and Its Mechanism in Humans and Mice. J. Immunol. 2010, 184, 5874–5884.
- Wang, Y.; Hasegawa, M.; Imamura, R.; Kinoshita, T.; Kondo, C.; Konaka, K.; Suda, T. PYNOD, a novel Apaf-1/CED4-like protein is an inhibitor of ASC and caspase-1. Int. Immunol. 2004, 16, 777–786.
- Joly, S.; Eisenbarth, S.C.; Olivier, A.K.; Williams, A.; Kaplan, D.H.; Cassel, S.L.; Flavell, R.A.; Sutterwala, F.S. Cutting Edge: Nlrp10 Is Essential for Protective Antifungal Adaptive Immunity against Candida albicans. J. Immunol. 2012, 189, 4713–4717.
- Eisenbarth, S.C.; Williams, A.; Colegio, O.R.; Meng, H.; Strowig, T.; Rongvaux, A.; Henao-Mejia, J.; Thaiss, C.A.; Joly, S.; Gonzalez, D.G.; et al. NLRP10 is a NOD-like receptor essential to initiate adaptive immunity by dendritic cells. Nature 2012, 484, 510–513.
- Damm, A.; Giebeler, N.; Zamek, J.; Zigrino, P.; Kufer, T.A. Epidermal NLRP10 contributes to contact hypersensitivity responses in mice. Eur. J. Immunol. 2016, 46, 1959–1969.
- Macaluso, F.; Nothnagel, M.; Parwez, Q.; Petrasch-Parwez, E.; Bechara, F.G.; Epplen, J.T.; Hoffjan, S. Polymorphisms in NACHT-LRR (NLR) genes in atopic dermatitis. Exp. Dermatol. 2007, 16, 692–698.
- Suárez-Fariñas, M.; Tintle, S.J.; Shemer, A.; Chiricozzi, A.; Nograles, K.; Cardinale, I.; Duan, S.; Bowcock, A.M.; Krueger, J.G.; Guttman-Yassky, E. Nonlesional atopic dermatitis skin is characterized by broad terminal differentiation defects and variable immune abnormalities. J. Allergy Clin. Immunol. 2011, 127, 954–964.e4.
- Reubold, T.F.; Hahne, G.; Wohlgemuth, S.; Eschenburg, S. Crystal structure of the leucine-rich repeat domain of the NOD-like receptor NLRP1: Implications for binding of muramyl dipeptide. FEBS Lett. 2014, 588, 3327–3332.
- Arnoult, D.; Soares, F.; Tattoli, I.; Castanier, C.; Philpott, D.J.; Girardin, S.E. An N-terminal addressing sequence targets NLRX1 to the mitochondrial matrix. J. Cell Sci. 2009, 122, 3161–3168.
- Moore, C.B.; Bergstralh, D.T.; Duncan, J.A.; Lei, Y.; Morrison, T.E.; Zimmermann, A.G.; Accavitti-Loper, M.A.; Madden, V.J.; Sun, L.; Ye, Z.; et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature 2008, 451, 573–577.
- Tattoli, I.; Carneiro, L.A.; Jéhanno, M.; Magalhaes, J.G.; Shu, Y.; Philpott, D.J.; Arnoult, D.; Girardin, S.E. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-κB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008, 9, 293–300.
- Hung, S.-C.; Huang, P.-R.; Almeida-da-Silva, C.L.C.; Atanasova, K.R.; Yilmaz, O.; Ojcius, D.M. NLRX1 modulates differentially NLRP3 inflammasome activation and NF-κB signaling during Fusobacterium nucleatum infection. Microbes Infect. 2018, 20, 615–625.
- Fekete, T.; Bencze, D.; Bíró, E.; Benkő, S.; Pázmándi, K. Focusing on the Cell Type Specific Regulatory Actions of NLRX1. Int. J. Mol. Sci. 2021, 22, 1316.




