
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Philippe Grange | + 2359 word(s) | 2359 | 2021-06-04 07:55:01 | | | |
| 2 | Nora Tang | Meta information modification | 2359 | 2021-06-25 09:36:47 | | | | |
| 3 | Conner Chen | Meta information modification | 2359 | 2021-09-22 04:32:14 | | |
Video Upload Options
Cutibacterium acnes is a member of the skin microbiota found predominantly in regions rich in sebaceous glands. It is involved in maintaining healthy skin and has long been considered a commensal bacterium. Its involvement in various infections has led to its emergence as an opportunist pathogen. Interactions between C. acnes and the human host, including the human skin microbiota, promote the selection of C. acnes strains capable of producing several virulence factors that increase inflammatory capability. This pathogenic property may be related to many infectious mechanisms, such as an ability to form biofilms and the expression of putative virulence factors capable of triggering host immune responses or enabling C. acnes to adapt to its environment. During the past decade, many studies have identified and characterized several putative virulence factors potentially involved in the pathogenicity of this bacterium. These virulence factors are involved in bacterial attachment to target cells, polysaccharide-based biofilm synthesis, molecular structures mediating inflammation, and the enzymatic degradation of host tissues. C. acnes, like other skin-associated bacteria, can colonize various ecological niches other than skin. It produces several proteins or glycoproteins that could be considered to be active virulence factors, enabling the bacterium to adapt to the lipophilic environment of the pilosebaceous unit of the skin, but also to the various organs it colonizes.
1. C. acnes Characteristics
- (1)
-
The genus Propionibacterium, comprising the species P. freundenreichii, P. cyclohexanicum, P. acidifaciens, and P. australianse.
- (2)
-
The new genus Acidipropionibacterium, comprising the species A. jensenii, A. thoenii, A. acidipropionici, A. microaerophilum, A. damnosum, and A. olivae.
- (3)
-
The new genus Pseudopropionibacterium, containing a single species: P. propionicum.
- (4)
-
The new genus Cutibacterium, comprising cutaneous Propionibacterium bacteria belonging to the species C. acnes, C. avidum, C. granulosum, and C. humerusii. P. acnes has thus been renamed C. acnes [3]. Moreover, the genus C. acnes has been further subdivided into subspecies, such as C. acnes subsp. defendens [4][5], and C. acnes subsp. elongatum [6] (Figure 1).
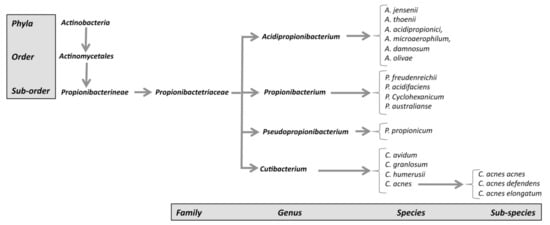 Figure 1. New classification of C. acnes strains.
Figure 1. New classification of C. acnes strains.
2. C. acnes Classification
| Phylotypes | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| C. acnes subspecies | WGS/Ribotyping a | SLST b | MLST8 c | CC | MLST9 d | CC | MALDI-TOF e | MLVA13 f | |
| C. acnes acnes | IA-1 | RT1, RT5 | A1-A34 | IA1 | CC1 | I-1a | CC18 | IA | IAI |
| RT532 | B1 | ||||||||
| IA-2 | RT1, RT4 | C1-C5 | IA1 | CC3 | I-1a | CC3 | IA | IA2 | |
| RT5 | |||||||||
| IB-1 | RT8 | D1-D5 | IA1 | CC4 | I-1a | CC28 | IB | IB | |
| E1-E9 | CC31 | ||||||||
| IB-2 | RT3, RT16 | F1-F14 | IA2 | CC2 | I-1b | CC28 | IB | ||
| IB-3 | RT1 | H1-H8 | IB | CC5 | I-2 | CC36 | IB | ||
| IC | RT5 | G1 | IC | CC107 | / | / | IB(IC) | / | |
| C. acnes defendens | RT2, RT6 | K1-K25 | CC6 | CC53 | |||||
| II | RT6 | II | CC30 | II | CC60 | II | II | ||
| CC71 | |||||||||
| CC72 | |||||||||
| C. acnes elongatum | III | RT9 | L1-L10 | III | CC77 | III | CC43 | III | III |
3. Skin Microbiota and Acne
References
- Gajdács, M.; Spengler, G.; Urbán, E. Identification and Antimicrobial Susceptibility Testing of Anaerobic Bacteria: Rubik’s Cube of Clinical Microbiology? Antibiotics 2017, 6, 25.
- Wickham, L. The Microbial Origin of Baldness: Sabouraud’s Researches into the Relations between Seborrhoea, Alopecia Areata, and Baldness. Br. Med. J. 1897, 1, 1028–1030.
- Scholz, C.F.P.; Kilian, M. The Natural History of Cutaneous Propionibacteria, and Reclassification of Selected Species within the Genus Propionibacterium to the Proposed Novel Genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 4422–4432.
- McDowell, A.; Barnard, E.; Liu, J.; Li, H.; Patrick, S. Proposal to Reclassify Propionibacterium acnes Type I as Propionibacterium acnes subsp. acnes subsp. nov. and Propionibacterium acnes Type II as Propionibacterium acnes subsp. defendens subsp. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5358–5365.
- McDowell, A.; Barnard, E.; Liu, J.; Li, H.; Patrick, S. Corrigendum: Proposal to Reclassify Propionibacterium acnes Type I as Propionibacterium acnes subsp. acnes subsp. nov. and Propionibacterium acnes Type II as Propionibacterium acnes subsp. defendens subsp. nov. Int. J. Syst. Evol. Microbiol. 2017, 67, 4880–4880.
- Dekio, I.; Culak, R.; Misra, R.; Gaulton, T.; Fang, M.; Sakamoto, M.; Ohkuma, M.; Oshima, K.; Hattori, M.; Klenk, H.-P.; et al. Dissecting the Taxonomic Heterogeneity within Propionibacterium acnes: Proposal for Propionibacterium acnes subsp. acnes subsp. nov. and Propionibacterium acnes subsp. elongatum subsp. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 4776–4787.
- Jeon, J.; Park, S.C.; Her, J.; Lee, J.W.; Han, J.-K.; Kim, Y.-K.; Kim, K.P.; Ban, C. Comparative Lipidomic Profiling of the Human Commensal Bacterium Propionibacterium acnes and Its Extracellular Vesicles. RSC Adv. 2018, 8, 15241–15247.
- Whale, G.A.; Sutcliffe, I.C.; Morrisson, A.R.; Pretswell, E.L.; Emmison, N. Purification and Characterisation of Lipoglycan Macroamphiphiles from Propionibacterium acnes. Antonie Van Leeuwenhoek 2004, 86, 77–85.
- Hall, G.S.; Pratt-Rippin, K.; Meisler, D.M.; Washington, J.A.; Roussel, T.J.; Miller, D. Growth Curve for Propionibacterium acnes. Curr. Eye Res. 1994, 13, 465–466.
- Davidsson, S.; Carlsson, J.; Mölling, P.; Gashi, N.; Andrén, O.; Andersson, S.-O.; Brzuszkiewicz, E.; Poehlein, A.; Al-Zeer, M.A.; Brinkmann, V.; et al. Prevalence of Flp Pili-Encoding Plasmids in Cutibacterium acnes Isolates Obtained from Prostatic Tissue. Front. Microbiol. 2017, 8, 2241.
- Dreno, B.; Thiboutot, D.; Gollnick, H.; Bettoli, V.; Kang, S.; Leyden, J.J.; Shalita, A.; Torres, V. Antibiotic Stewardship in Dermatology: Limiting Antibiotic Use in Acne. Eur. J. Dermatol. 2014, 24, 330–334.
- Johnson, J.L.; Cummins, C.S. Cell Wall Composition and Deoxyribonucleic Acid Similarities among the Anaerobic Coryneforms, Classical Propionibacteria, and Strains of Arachnia propionica. J. Bacteriol. 1972, 109, 1047–1066.
- Webster’, G.F.; Cummins, C.S. Use of Bacteriophage Typing to Distinguish Propionibacterium acnes Types I and II. J. Clin. Microbiol. 1978, 7, 84–90.
- Perry, A.L.; Worthington, T.; Hilton, A.C.; Lambert, P.A.; Stirling, A.J.; Elliott, T.S.J. Analysis of Clinical Isolates of Propionibacterium acnes by Optimised RAPD. FEMS Microbiol. Lett. 2003, 228, 51–55.
- McDowell, A.; Valanne, S.; Ramage, G.; Tunney, M.M.; Glenn, J.V.; McLorinan, G.C.; Bhatia, A.; Maisonneuve, J.-F.; Lodes, M.; Persing, D.H.; et al. Propionibacterium acnes Types I and II Represent Phylogenetically Distinct Groups. J. Clin. Microbiol. 2005, 43, 9.
- McDowell, A.; Perry, A.L.; Lambert, P.A.; Patrick, S. A New Phylogenetic Group of Propionibacterium acnes. J. Med Microbiol. 2008, 57, 218–224.
- Fitz-Gibbon, S.; Tomida, S.; Chiu, B.-H.; Nguyen, L.; Du, C.; Liu, M.; Elashoff, D.; Erfe, M.C.; Loncaric, A.; Kim, J.; et al. Propionibacterium acnes Strain Populations in the Human Skin Microbiome Associated with Acne. J. Investig. Dermatol. 2013, 133, 2152–2160.
- McDowell, A.; Nagy, I.; Magyari, M.; Barnard, E.; Patrick, S. The Opportunistic Pathogen Propionibacterium acnes: Insights into Typing, Human Disease, Clonal Diversification and CAMP Factor Evolution. PLoS ONE 2013, 8, e70897.
- McDowell, A.; Gao, A.; Barnard, E.; Fink, C.; Murray, P.I.; Dowson, C.G.; Nagy, I.; Lambert, P.A.; Patrick, S. A Novel Multilocus Sequence Typing Scheme for the Opportunistic Pathogen Propionibacterium acnes and Characterization of Type I Cell Surface-Associated Antigens. Microbiology 2011, 157, 1990–2003.
- McDowell, A.; Barnard, E.; Nagy, I.; Gao, A.; Tomida, S.; Li, H.; Eady, A.; Cove, J.; Nord, C.E.; Patrick, S. An Expanded Multilocus Sequence Typing Scheme for Propionibacterium acnes: Investigation of ‘Pathogenic’, ‘Commensal’ and Antibiotic Resistant Strains. PLoS ONE 2012, 7, e41480.
- Barnard, E.; Nagy, I.; Hunyadkürti, J.; Patrick, S.; McDowell, A. Multiplex Touchdown PCR for Rapid Typing of the Opportunistic Pathogen Propionibacterium acnes. J. Clin. Microbiol. 2015, 53, 1149–1155.
- Lomholt, H.B.; Kilian, M. Population Genetic Analysis of Propionibacterium acnes Identifies a Subpopulation and Epidemic Clones Associated with Acne. PLoS ONE 2010, 5, e12277.
- Kilian, M.; Scholz, C.F.P.; Lomholt, H.B. Multilocus Sequence Typing and Phylogenetic Analysis of Propionibacterium acnes. J. Clin. Microbiol. 2012, 50, 1158–1165.
- Scholz, C.F.P.; Jensen, A.; Lomholt, H.B.; Brüggemann, H.; Kilian, M. A Novel High-Resolution Single Locus Sequence Typing Scheme for Mixed Populations of Propionibacterium acnes in Vivo. PLoS ONE 2014, 9, e104199.
- Paugam, C.; Corvec, S.; Saint-Jean, M.; Le Moigne, M.; Khammari, A.; Boisrobert, A.; Nguyen, J.M.; Gaultier, A.; Dréno, B. Propionibacterium acnes Phylotypes and Acne Severity: An Observational Prospective Study. J. Eur. Acad. Dermatol. Venereol. 2017, 31, e398–e399.
- Broly, M.; Ruffier d’Epenoux, L.; Guillouzouic, A.; Le Gargasson, G.; Juvin, M.-E.; Leroy, A.G.; Bémer, P.; Corvec, S. Propionibacterium/Cutibacterium Species–Related Positive Samples, Identification, Clinical and Resistance Features: A 10-Year Survey in a French Hospital. Eur J. Clin. Microbiol. Infect. Dis 2020, 39, 1357–1364.
- Teramoto, K.; Okubo, T.; Yamada, Y.; Sekiya, S.; Iwamoto, S.; Tanaka, K. Classification of Cutibacterium acnes at Phylotype Level by MALDI-MS Proteotyping. Proc. Jpn. Acad. Ser. B 2019, 95, 612–623.
- Hauck, Y.; Soler, C.; Gérôme, P.; Vong, R.; Macnab, C.; Appere, G.; Vergnaud, G.; Pourcel, C. A Novel Multiple Locus Variable Number of Tandem Repeat (VNTR) Analysis (MLVA) Method for Propionibacterium acnes. Infect. Genet. Evol. 2015, 33, 233–241.
- Tomida, S.; Nguyen, L.; Chiu, B.-H.; Liu, J.; Sodergren, E.; Weinstock, G.M.; Li, H. Pan-Genome and Comparative Genome Analyses of Propionibacterium acnes Reveal Its Genomic Diversity in the Healthy and Diseased Human Skin Microbiome. mBio 2013, 4, e00003-13.
- Dagnelie, M.-A.; Khammari, A.; Dréno, B.; Corvec, S. Cutibacterium acnes Molecular Typing: Time to Standardize the Method. Clin. Microbiol. Infect. 2018, 24, 1149–1155.
- Nagy, E.; Urbán, E.; Becker, S.; Kostrzewa, M.; Vörös, A.; Hunyadkürti, J.; Nagy, I. MALDI-TOF MS Fingerprinting Facilitates Rapid Discrimination of Phylotypes I, II and III of Propionibacterium acnes. Anaerobe 2013, 20, 20–26.
- Belkaid, Y.; Segre, J.A. Dialogue between Skin Microbiota and Immunity. Science 2014, 346, 954–959.
- Grice, E.A. The Intersection of Microbiome and Host at the Skin Interface: Genomic- and Metagenomic-Based Insights. Genome Res. 2015, 25, 1514–1520.
- Borkowski, A.W.; Gallo, R.L. The Coordinated Response of the Physical and Antimicrobial Peptide Barriers of the Skin. J. Investig. Dermatol. 2011, 131, 285–287.
- Svensson, A.; Ofenloch, R.F.; Bruze, M.; Naldi, L.; Cazzaniga, S.; Elsner, P.; Goncalo, M.; Schuttelaar, M.-L.A.; Diepgen, T.L. Prevalence of Skin Disease in a Population-Based Sample of Adults from Five European Countries. Br. J. Dermatol. 2018, 178, 1111–1118.
- Cotterill, J.A.; Cunliffe, W.J. Suicide in Dermatological Patients. Br. J. Dermatol. 1997, 137, 246–250.
- Zaenglein, A.L. Acne Vulgaris. N. Engl. J. Med. 2018, 379, 1343–1352.
- Bocquet-Trémoureux, S.; Corvec, S.; Khammari, A.; Dagnelie, M.-A.; Boisrobert, A.; Dreno, B. Acne Fulminans and Cutibacterium acnes Phylotypes. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 827–833.
- Stewart, M.E.; Steele, W.A.; Downing, D.T. Changes in the Relative Amounts of Endogenous and Exogenous Fatty Acids in Sebaceous Lipids during Early Adolescence. J. Invest. Dematol. 1989, 92, 371–378.
- Deplewski, D.; Rosenfield, R.L. Role of Hormones in Pilosebaceous Unit Development. Endocr. Rev. 2000, 21, 30.
- Tax, G.; Urbán, E.; Palotás, Z.; Puskás, R.; Kónya, Z.; Bíró, T.; Kemény, L.; Szabó, K. Propionic Acid Produced by Propionibacterium acnes Strains Contributes to Their Pathogenicity. Acta Dermatol. Venerol. 2016, 96, 43–49.
- Hall, J.B.; Cong, Z.; Imamura-Kawasawa, Y.; Kidd, B.A.; Dudley, J.T.; Thiboutot, D.M.; Nelson, A.M. Isolation and Identification of the Follicular Microbiome:IImplications for Acne Research. J. Investig. Dermatol. 2018, 138, 2033–2040.
- Prast-Nielsen, S.; Tobin, A.-M.; Adamzik, K.; Powles, A.; Hugerth, L.W.; Sweeney, C.; Kirby, B.; Engstrand, L.; Fry, L. Investigation of the Skin Microbiome: Swabs vs. Biopsies. Br. J. Dermatol. 2019, 181, 572–579.
- Dreno, B.; Martin, R.; Moyal, D.; Henley, J.B.; Khammari, A.; Seité, S. Skin Microbiome and Acne Vulgaris: Staphylococcus, a New Actor in Acne. Exp. Dermatol. 2017, 26, 798–803.
- Grice, E.A.; Segre, J.A. The Skin Microbiome. Nat. Rev Microbiol. 2011, 9, 244–253.
- Nakase, K.; Hayashi, N.; Akiyama, Y.; Aoki, S.; Noguchi, N. Antimicrobial Susceptibility and Phylogenetic Analysis of Propionibacterium acnes Isolated from Acne Patients in Japan between 2013 and 2015. J. Dermatol. 2017, 44, 1248–1254.
- Johnson, T.; Kang, D.; Barnard, E.; Li, H. Strain-Level Differences in Porphyrin Production and Regulation in Propionibacterium acnes Elucidate Disease Associations. mSphere 2016, 1, e00023-15.
- Dréno, B.; Pécastaings, S.; Corvec, S.; Veraldi, S.; Khammari, A.; Roques, C. Cutibacterium acnes (Propionibacterium acnes) and Acne Vulgaris: A Brief Look at the Latest Updates. J. Eur. Acad. Dermatol. Venereol. 2018, 32, 5–14.
- Xu, H.; Li, H. Acne, the Skin Microbiome, and Antibiotic Treatment. Am. J. Clin. Dermatol. 2019, 20, 335–344.
- Barnard, E.; Shi, B.; Kang, D.; Craft, N.; Li, H. The Balance of Metagenomic Elements Shapes the Skin Microbiome in Acne and Health. Sci. Rep. 2016, 6, 39491.
- Meisel, J.S.; Sfyroera, G.; Bartow-McKenney, C.; Gimblet, C.; Bugayev, J.; Horwinski, J.; Kim, B.; Brestoff, J.R.; Tyldsley, A.S.; Zheng, Q.; et al. Commensal Microbiota Modulate Gene Expression in the Skin. Microbiome 2018, 6, 20.
- Paetzold, B.; Willis, J.R.; Pereira de Lima, J.; Knödlseder, N.; Brüggemann, H.; Quist, S.R.; Gabaldón, T.; Güell, M. Skin Microbiome Modulation Induced by Probiotic Solutions. Microbiome 2019, 7, 95.




