Proteins and DNA exhibit key physical chemical properties that make them advantageous for building nanostructures with outstanding features. Both DNA and protein nanotechnology have growth notably and proved to be fertile disciplines. The combination of both types of nanotechnologies is helpful to overcome the individual weaknesses and limitations of each one, paving the way for the continuing diversification of structural nanotechnologies. Recent studies have implemented a synergistic combination of both biomolecules to assemble unique and sophisticate protein–DNA nanostructures. These hybrid nanostructures are highly programmable and display remarkable features that create new opportunities to build on the nanoscale.
1. Introduction
The versatility and programmability of DNA on the nanoscale has been demonstrated by the notable growth and diversification that structural DNA nanotechnology has displayed in the last decades
[1][2][3][4]. The continues growth and diversification of structural DNA nanotechnology has been paved by the establishment of novel strategies to build DNA structures
[3][5]. For example, the building of DNA-junctions and DNA-crossovers
[1], pioneered by Seeman in the 1980s, and DNA origami
[6], developed by Rothemund in the early 2000s, were important milestones. These building blocks served as the foundation to build novel, complex, and hierarchical nanostructures that inaugurated new subfields in the genealogy of structural DNA nanotechnology
[3].
The scope of structural DNA nanotechnology has been further expanded by the incorporation of other type of building blocks
[3]. For example, the incorporation of inorganic nanoparticles by Mirkin and his collaborators lead to the development of programmable DNA-based colloidal crystals (commonly referred as spherical nucleic acids) with applications in photonics, electronics, and self-assembly
[5]. These hybrid nanomaterials combine ssDNA molecules with rigid templates made up of inorganic nanoparticles. The latter acts as the brick that organizes and dictates the shape, while the former works as “glue” by establishing directional “bonds”. DNA-based colloidal crystals are an early example of how the combination of DNA with other types of building blocks could generate a whole new area of programmable hybrid nanomaterials.
Similarly, the incorporation of proteins into structural DNA nanotechnology as a cobuilding block further expanded the scope of DNA nanotechnology and sprouted new research avenues
[7][8]. The use of proteins in DNA nanotechnology is indeed not new. Proteins have been implemented since the dawn of structural DNA nanotechnology
[9][10]. Nevertheless, the accumulated advances in the understanding of protein self-assembly, design, and engineering has increased interest in integrating them into DNA nanotechnology. However, until now, the incorporation of proteins has mostly been limited to equipping DNA nanostructures with specific functionalities, for example, molecular recognition
[11] or catalytic activity
[12]. This limited use of proteins contrast with the myriad functions and capabilities that nucleoprotein complexes perform in nature (e.g., genetic switches, ribosomes, nucleosomes, and viruses). Looking at the large structural and functional diversity of nucleoproteins, we can appreciate the potential that proteins have in terms of working synergically with DNA building blocks.
More recently, the incorporation of proteins to control or enhance the structural features or properties of DNA nanostructures has been implemented; however, it still remains largely unexplored. This review focuses on the structural roles that proteins offer for building hybrid nucleoprotein nanostructures through their combined self-assembly with diverse DNA building blocks (e.g., DNA origami, DNA junctions, plasmid, linear DNA). We review seminal and recent work to show the strategies deployed to build hybrid protein–DNA nanostructures. We demonstrate how the full integration of proteins into DNA nanotechnology, mainly through structural and mechanical roles, makes it possible to build remarkable and unique nanomaterials and exploit all the potential benefits that these hybrid materials can offer. In order to review the application of the nonstructural roles of proteins in DNA nanostructures (e.g., the arrangement of proteins or enzymes on preassembled DNA nanostructures) as well as the structural roles that DNA offers in building nanostructures, we suggest exploring other recently published reviews
[4][7][8][13].
2. Structural Protein–DNA Nanotechnology
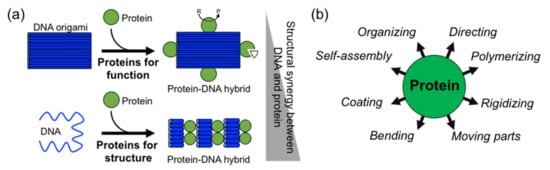
In comparison to DNA, proteins display more complex, sophisticate, and diverse functions, as well as a larger structural diversity. These include, for example, highly specific catalytic activity, potent molecular recognition, tight and precise allosteric regulation, efficient cargo encapsulation, responsive structural functions, and cooperative binding. Since the early developments in structural DNA nanotechnology, these functionalities have been harnessed to increase the functionality of DNA nanomaterials. They provide advanced functionalities such as enhanced recognition for cellular ligands or enzymatic cascades. To achieve this, proteins are precisely positioned on a previously assembled DNA nanostructure (e.g., DNA origami or DNA junctions) (a)
[9][10][11][14][15]. Moreover, by adding functional capabilities to otherwise inert DNA nanostructures, proteins can cooperate synergistically and bring important features to the final assembled hybrid nanostructure (a,b).
Figure 1. Overview of structural protein–DNA nanotechnology. (a) “Proteins for function” versus “proteins for structure” in DNA nanotechnology. In the latter case, there is much more synergy between the protein and the DNA building blocks. “R” means reactive and “P” product. (b) Structural roles of proteins in hybrid protein–DNA nanotechnology.
Two distinctive approaches to how proteins are combined with DNA can be clearly distinguished: (1) proteins for function and (2) proteins for structure (a). The first represents “functional Protein DNA nanotechnology”, whereas the second “structural Protein-DNA nanotechnology”. This review focuses on the second approach, which we refer to in the review as “structural protein-DNA nanotechnology”. In this hybrid protein–DNA nanotechnology, proteins and DNA act synergistically during the self-assembly process and serve as the foundation for the final nanostructure. Meaning that protein and DNA nanotechnologies show a high degree of structural integration.
By advantageously harnessing the biophysical and chemical properties from both biomolecules
[16], structural protein–DNA nanotechnology has reduced the limitations that each molecule present when used alone. Proteins have a larger chemical and structural diversity than DNA and, although proteins alone can build sophisticated nanostructures, their versatility and programmability are severely limited due to intricate sequence–structure relationships. On the other hand, DNA lacks the ample structural and chemical diversity seen in proteins but has more predictable folding than proteins due to the readily programmable Watson–Crick interactions. Since protein–DNA nanotechnology aims to harness the different but highly complementary physical–chemical and structural properties of both biomolecules, their synergistic combination offers strategical benefits for the fabrication of nanomaterials.
Structural protein–DNA nanotechnology is different from other common uses of proteins in DNA nanostructures because proteins play important structural, mechanical, and/or assembling roles. Although both DNA and proteins provide these roles, their degree of participation depends on the structural complexity of the starting and final structure. However, as it is shown below, most of the literature shows that the use of proteins is more operative than DNA. It is considered that proteins and DNA have structural roles in a particular protein–DNA nanostructure when it is not possible to achieve such a final nanostructure without the coparticipation of both building blocks (a). Hence, the nonexistence of one building block does not lead to the acquisition of a particular shape, size, order, organization, or certain mechanical or dynamic properties. This means that the removal or the absence of one of them (protein or DNA) disassembles the structure or largely compromises its stability or properties.
As a result of their large structural and chemical diversity, proteins can bring multiple advantages when used for structural purposes (b). They can spatially align DNA in specific geometries and preserve DNA topology by coating and stiffening. Furthermore, proteins can establish strong and specific interactions with ssDNA, and in particular, with DNA duplexes. This opens the opportunity to incorporate dsDNA into current DNA nanotechnology, which in turn relies on ssDNA (M13 virus plasmid and staple oligonucleotides)
[17][18]. As proteins offer the advantage of working isothermally and at environmental temperatures, they can reduce the dependence on DNA molecules and multitemperature assembly processes of DNA nanotechnology. Therefore, proteins have a large potential to significantly reduce the production costs and simplify assembly processes, which currently limits the large-scale use of DNA nanotechnology in many applications.
3. Proteins in Hybrid Nanotechnology
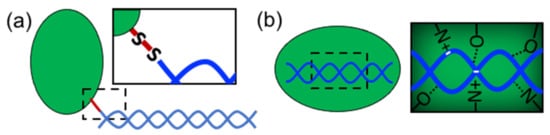
In order to form nanostructures with DNA, proteins need to establish strong and effective interactions with DNA building blocks. Several types of proteins have been used in structural hybrid nanotechnology. These include enzymes, multimeric proteins, metal-binding proteins, coiled-coil peptides, cationic peptides, cationic polymer proteins, ribosomal proteins, transcription factors, viral proteins, nucleosomes, polymerases, and others (). These proteins interact with DNA through two different approaches: (1) covalent conjugation or (2) noncovalent coassembly (). Covalent conjugation is usually performed by chemically linking proteins and DNA through reactive groups (a). The DNA can be an assembled nanostructure or oligos with complementary sequences. Covalent conjugation is frequently used because it is straightforward, and it is easy to control and render (bio)chemically stable conjugates
[13]. This strategy also means that practically any protein carrying the proper reactive group can be conjugated. On the other hand, noncovalent coassembly requires using proteins with DNA-binding capabilities (b). Since noncovalent interactions are tunable and reversible, they offer the possibility to create flexible, modular, and highly dynamic hybrid nanostructures with advanced and complex functionalities that can mimic natural nucleoprotein complexes. However, the resultant complexes can have low stability and be more susceptible to the environmental conditions than chemically linked complexes; thus, the control of these types of interaction represents a great challenge.
Figure 2. Approaches to link proteins and DNA. (a) Covalent conjugation (“S-S” represents a disulfide bridge) and (b) noncovalent interactions (dotted lines represent hydrogen bonds).
The reported proteins (lacking DNA-binding capabilities) conjugated covalently to DNA include (multimeric) enzymes used as structural templates
[19][20][21], metal-binding proteins
[22], coiled-coil peptides
[23], and elastine-like peptides
[24]. By contrast, proteins that exhibit a DNA binding affinity in a sequence-dependent or independent mode include cationic peptides
[25][26][27][28], cationic polymer proteins
[29][30][31], ribosomal proteins
[32], transcription activator-like (TAL) effectors
[17], transcription factors
[33], viral proteins
[34][35][36], histones, and polymerases
[37]. Simpler options such as streptavidin or bioinspired cationic protein polymers made up of extremely simple and repetitive amino acids that retain DNA-binding functionality or even virus-like properties have also been used
[7][25][38]. These proteins can be used in combination with junctions, tiles, motifs, or origamis, and also single ssDNA or dsDNA molecules.
Table 1. Proteins used in structural protein–DNA nanotechnology.
| Building Block |
Type of Protein |
Building Strategy |
Interaction 1 |
Ref. |
| βGalactoside 1D-DNA conjugate |
Enzyme |
(1) Structural scaffold to attach DNA |
C |
[19] |
| (2) Polymerization |
| GroEL-DNA conjugate |
Chaperonin |
(1) Structural scaffold to attach DNA |
C |
[20] |
| (2) Polymerization |
| RIDC3-DNA conjugate |
Engineered tetrameric metal-interacting cytochrome cb56 |
(1) Structural scaffold to attach DNA |
C |
[22] |
| (2) Polymerization |
| Drosophila Engrailed homeodomain (ENH) |
Engineered transcription factor |
(1) Polymerization |
NC |
[33] |
| Coiled coil-DNA conjugate |
De novo dimerizing peptide |
(1) Polymerization |
C |
[23] |
| K3C6SPD |
Engineered self-assembly β-sheet cationic peptide |
(1) Polymerization |
NC |
[25] |
| CP++ and sCP |
Designed self-assembly cationic collagen mimetic peptides |
(1) Polymerization |
NC |
[27] |
| Aldolase-DNA conjugate |
Trimeric enzyme |
(1) Structural scaffold to attach DNA |
C |
[21] |
| (2) Spatial organization |
| H2A, H2B, H3 and H4 |
Histone proteins forming nucleosomes (Chromatin) |
(1) Spatial organization |
NC |
[39] |
| Streptavidin |
Tetrameric biotin binding protein |
(1) Spatial organization |
NC |
[40][41] |
| Traptavidin |
Engineered tetrameric biotin binding protein |
(1) Spatial organization |
NC |
[42] |
| I3V3A3G3K3 |
Engineered self-assembly β-sheet cationic peptide |
(1) No programmable folding of DNA |
NC |
[28] |
| L7Ae |
RNA-binding ribosomal protein |
(1) Bending |
NC |
[32][43] |
| (2) Conformational change |
| Transcription activator–like (TAL) effector |
Engineered bivalent proteins for recognition of specific DNA sequences |
(1) Programmable folding of DNA |
NC |
[17] |
| RecA |
DNA-binding protein involved in the repair and maintenance of DNA |
(1) Self-assembly |
NC |
[44] |
| (2) Coating |
| (3) Rigidifying |
| Tobacco Mosaic Virus coat protein |
Viral RNA binding protein |
(1) Self-assembly |
NC |
[34][35] |
| (2) Coating |
| (3) Rigidifying |
| (4) Dynamic systems |
| Redβ |
Single-strand annealing protein for homologous recombination in phages |
(1) Coating |
NC |
[36] |
| (2) Rigidifying |
| C8-BSso7d |
Engineered diblock protein polymer carrying a nonsequence specific dsDNA binding domain from archeal origin |
(1) Coating |
NC |
[18][31][45] |
| (2) Rigidifying |
| C4-S10-BK12 |
Engineered triblock cationic protein polymer |
(1) Coating |
NC |
[30] |
| (2) Rigidifying |
| C4-BK12 |
Engineered diblock cationic protein polymer |
(1) Coating |
NC |
[29][46] |
| (2) Rigidifying |
| T7RNAP-ZIF |
Engineered T7 RNA polymerase fused to a DNA-binding zinc finger motif |
(1) Moving DNA parts |
NC |
[37] |
| (GVGVP)40 |
Engineered elastin-like polypeptide |
(1) Dynamic and responsive systems |
C |
[24] |
4. Strategies to Build Protein–DNA Nanostructures
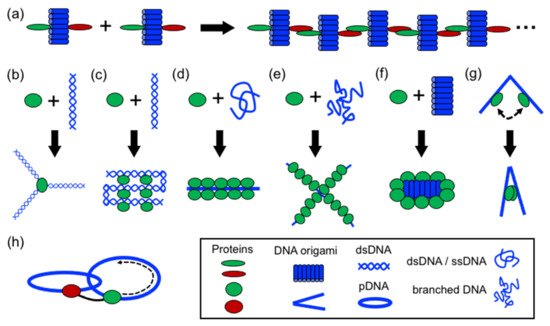
In structural protein–DNA nanotechnology, proteins and DNA building blocks have been combined following a variety of strategies. The strategies reported until now include polymerization, directing and spatial organization, bending, folding, self-assembly, coating, rigidizing, and moving DNA parts (). In these processes, DNA and proteins act synergically to build more complex structures. However, proteins generally have a more active role during the assembly than DNA. However, DNA plays an important role in the self-assembly of the final structure by operating as a structural template or scaffold for protein binding or anchoring. Below, it is discussed the aforementioned strategies.
Figure 3. Strategies in structural protein–DNA nanotechnology. (a) Polymerizing; (b) directing spatial organization; (c) shaping DNA through bending and folding; (d) protein self-assembly on DNA; (e,f) coating and rigidizing DNA; (g) switching and (h) moving DNA components. DNA origami, ssDNA, and dsDNA molecules are depicted in blue; proteins are depicted in green and red.
5. Functional Applications
A large and diverse list of potential applications for this relatively new type of nanomaterial has been suggested. However, instead of demonstrating their applications, researchers put considerable effort into establishing basic rules and general guidelines for fabrication. Indeed, most applications of protein–DNA nanostructures are currently in the “proof-of-concept” stage of investigation. Another interesting point is that many researchers envision their hybrid nanostructures as versatile platforms for many different applications.
The most anticipated applications are in the fields of nanomedicine, synthetic biology, structural biology and biophysics, bioinspired nanomaterials, and nanorobotics. Within nanomedicine, the development of nanobiosensors for molecular imaging, as well as smart and responsive drug and gene delivery systems are being explored
[43]. The latter is the most studied application
[38][47][48]. The combination of sensing and drug delivery could lead to the creation of theragnostic hybrid materials
[42]. Other applications of high relevance for nanomedicine are the creation of bioresponsive nanomaterials
[24] and immunomaterials, such as multivalent vaccines. Potential applications inside structural biology and biophysics include the establishment of nanoplatforms for investigating the structure and assembly mechanisms of viruses
[30][49] and controlling the architecture of genomic DNA and gene expression by looping DNA
[17]. Protein–DNA nanostructures acting as scaffolds to attach multiple enzymes involved in the biosynthetic pathways
[50] of high-value molecules are of particular interest in synthetic biology.
An area in which hybrid protein–DNA nanostructures have a particularly large potential is the mimicking of nucleoprotein complexes such as viruses, transcription factors, and ribosomes. Viromimetic hybrid materials have been developed into gene delivery systems and simple virus-like models for biophysical and structural studies
[25][28][30][49]. With the development of dynamic protein–DNA nanostructures
[37], it is possible to envision mimicking more complex structures such as ribosomes or motors for injecting DNA
[35]. More advanced potential applications involve the fabrication of nanodevices, smart machines, and nanorobots. These molecular nanomachines could detect signals, localize target proteins, and control living cell function and fate
[51]. Finally, nonbiological applications include the development of materials with catalytic properties, nanowires for nanoelectronics, and broader nature-inspired nanotechnology.