
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Diego Franco | + 1723 word(s) | 1723 | 2021-02-19 10:13:11 | | | |
| 2 | Vicky Zhou | Meta information modification | 1723 | 2021-02-26 06:10:12 | | |
Video Upload Options
Cardiac development is a complex developmental process that is initiated soon after gastrulation, as two sets of precardiac mesodermal precursors are symmetrically located and subsequently fused at the embryonic midline forming the cardiac straight tube. Thereafter, the cardiac straight tube invariably bends to the right, configuring the first sign of morphological left–right asymmetry and soon thereafter the atrial and ventricular chambers are formed, expanded and progressively septated. As a consequence of all these morphogenetic processes, the fetal heart acquired a four-chambered structure having distinct inlet and outlet connections and a specialized conduction system capable of directing the electrical impulse within the fully formed heart.
1. Introduction
Over the last decades, our understanding of the cellular and molecular mechanisms driving cardiac development has greatly increased. Such discoveries have provided clues to dissect the genetic and molecular bases of congenital heart diseases, as well as provided tools to configure novel cellular and molecular approaches to heal the damaged heart. In this review, we provide a comprehensive summary of the cellular and molecular pathways involved in heart formation, and translational contribution of such findings.
2. From Gastrulation to the Early Cardiac Linear Tube
Cardiac development is complex developmental process that is initiated soon after gastrulation. Soon thereafter the epiblast starts delaminating and migrating towards the future mesodermal layer, and cardiac precursors can be traced in the primitive streak (Figure 1A). At this stage, mesoderm precursors are characterized by the expression of Brachyury [1], Mesp1 and Mesp2 [2][3][4], having all of them an important contribution to early cardiogenic development [5][6][7][8][9]. Following the first configuration of the mesodermal layer, cardiac precursors migrate anteriorly and they start expressing early cardiogenic transcription factors. At this stage, several members of the Forkhead, Nkx, Gata, and Mef2 families, respectively, are expressed in the precardiac mesoderm [10][11][12][13] in a wide range of different species such as Xenopus [14], zebrafish [15][16][17], chicken [18][19][20][21][22], and mice [23][24][25][26][27][28][29][30], being particularly important Nkx2.5 [31][32][33][34], Gata4 [35][36][37][38], and Mef2c [39][40][41][42][43] for early cardiogenesis in different experimental models. Expression of these cardiogenic markers is regulated by signaling factors from the adjacent tissues such as Bmp2 [44][45][46][47][48][49], Fgf8 [50][51][52][53] and Wnt signaling ][54][55][56][57][58][59][60]. Importantly, Bmp2 and Fgf8 signaling is controlled by an intricated molecular cascade in which non-coding RNAs, such as miR-130 and miR-133, are also involved [61]. As development proceed, the initial cardiogenic precursors become configured into a horseshoe shape (Figure 1B) and subsequently the bilateral cardiogenic precursors are fused in the embryonic midline generating a cardiac straight tube [62]. At this stage, the embryonic heart is configured by two distinct epithelial layers, an externally located myocardial layer and an internally located endocardial layer. The two distinct cardiogenic subpopulations can be already recognized at this stage [63][64], the first heart field (FHF) that contributes to the early cardiac straight tube, and the second heart field (SHF) that is medially located and will subsequently contribute to both the arterial and venous poles of the heart [65][66][67] (Figure 1C). FHF specification is dependent of cardiogenic factors such as Nkx2.5 [68], while SHF specification is mostly determined by islet-1 [69] while Bmp signaling contributes to SHF proliferation [70]. On the other hand, Mef2c is required for both FHF and SHF development [71].
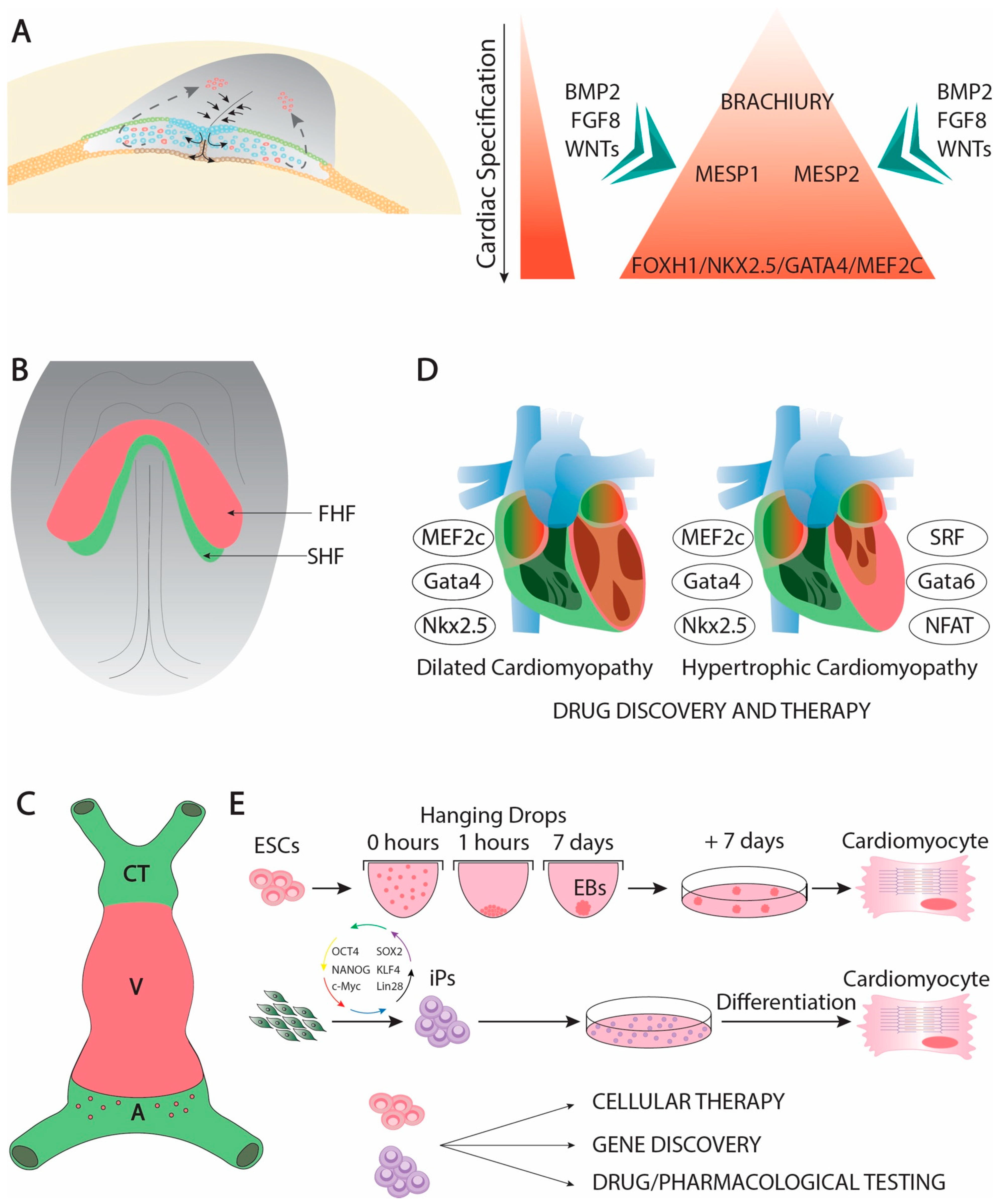
Genetically modified mice have uncovered key functional roles of distinct transcription factors during early cardiac developmental stages. In this context, Gata4 systemic mutants leads to absence of cardiac tube formation [94][95]. Mef2c and Foxh1 systemic mutants are arrested at the cardiac linear heart tube stage [28][35], while Nkx2.5 mutants failed to develop right after cardiac looping, providing some cues for ventricular left–right development [96]. Importantly, Mef2c, Gata4, and Nkx2.5 constitute coregulators of early cardiac development [97][98][99][100]. Moreover, these core cardiac transcription factors can also be associated with additional cofactors during early cardiogenesis such as Tbx5 [101][102][103][104], Gata5 [105], and Gata 6 [106], constituting a complex gene regulatory network [107][108][109] that also implies other transcription factors as reported in different experimental models [110][111][112][113][114].
These transcriptional networks have been studied in detail over the last decades, uncovering a multiple of downstream genes [115][116][117][118][119][120][121][122], including non-coding RNAs, such as miR-99/let-7 [123][124], that are involved in multiple steps of cardiac development as reported in different species. Overall, these data demonstrate that early cardiac development is an intricated morphogenetic mechanisms in which multiple factors are critically involved (Table 1).
Table 1. List of distinct transcription factors involved in cardiogenesis and they main functional contribution to heart development.
| TF | Function | References |
|---|---|---|
| Brachyury | Mesodermal commitment | [1] |
| Mesp1 | Cardiogenic mesoderm commitment | [2][3][4][5][6][7][8][9] |
| Mesp2 | Cardiogenic mesoderm commitment | [2][3][4][5][6][7][8][9] |
| Gata4 | Early cardiac specification, proepicardium development, chamber formation, atrial and atrioventricular septation, cardiomyocyte proliferation, cardiac hypertrophy | [10][11][12][13][14][27][35][36][37][38][94][95][97][98][99][100][125][126][127] |
| Gata5 | Early cardiac specification, cardiomyocyte proliferation | [10][11][12][13][14][16][26][105] |
| Gata6 | Early cardiac specification, cardiac hypertrophy | [10][11][12][13][14][16][25][41] |
| Nkx2.3 | Early cardiac specification | [15] |
| Nkx2.5 | Early cardiac specification, FHF development, cardiac looping, chamber formation, cardiac conduction system specification, atrial septation | [15][31][32][33][34][68][96][97][98][99][100][128][129][130][131][132][133][134][135] |
| Nkx2.6 | Early cardiac specification | [24][29] |
| Nkx2.7 | Early cardiac specification | [15] |
| Nkx2.8 | Early cardiac specification | [18][19][20] |
| Foxh1 | Anterior heart field development | [28][35] |
| Mef2c | Early cardiac specification, chamber formation | [35][36][37][38][39][40][41][42][43][136] |
| Islet-1 | Second heart field specification | [69] |
| Tbx5 | Early cardiac specification, chamber formation, cardiac conduction system specification, atrial and ventricular septation | [101][102][103][104][128][137][138][139][140][141][142] |
| Pitx2 | left right signalling, heterotaxia, chamber formation, cardiac conduction system development | [143][144][145][146][147][148][149][150][151][152][153][154][155][156][157][158][159][160][161][162][163][164][165][166][167] |
| Prrx1 | cardiac looping | [168] |
| Wt1 | proepicardium and epicardial development | [169][170] |
| Tcf21 | proepicardium and epicardial development | [171][172] |
| Tbx18 | proepicardium and epicardial development | [169] |
| Tbx2 | primary myocardium and cardiac conduction system development | [173][174][175][176][177][178] |
| Tbx3 | primary myocardium and cardiac conduction system development | [179][180][181][182] |
| Tbx20 | chamber formation, atrioventricular canal development, cardiomyocyte cell proliferation | [183][184][185][186][187][188][189][190] |
| Irx3 | ventricular trabecular and cardiac conduction system development | [191][192] |
| Irx4 | ventricular chamber development | [191][193] |
| Irx5 | cardiac conduction system development | [191][192] |
| Hey1 | ventricular chamber development | [194][195][196] |
| Hey2 | ventricular chamber development | [194][195][196][197] |
| Coup-TFII | atrial development | [198] |
| eHand | ventricular development | [199][200][201][202] |
| dHand | ventricular development | [199][202] |
| Foxm1 | ventricular trabecular and compact layers development | [203] |
| Hop | ventricular trabecular and compact layers development | [204] |
| Klf13 | ventricular trabecular and compact layers development | [205] |
| Srf | ventricular trabecular and compact layers development | [206][207] |
| Shox2 | cardiac conduction development | [208][209][210][211][212][213] |
| Odd1 | atrial septation | [214] |
| Klf2 | atrioventricular septation | [215] |
| Sox9 | atrioventricular septation | [216] |
| Smad4 | atrioventricular septation | [217] |
| Tbx1 | outflow tract and aortic arch development | [218] |
| Foxc1 | aortic arch development | [219] |
| Foxc2 | aortic arch development | [219] |
| Prx1 | aortic arch development | [220] |
| Prx2 | aortic arch development | [220] |
3. Perspectives
Over the last decades, our understanding of the cellular and molecular processes involved in cardiac morphogenesis has greatly increased. A large number of studied in different species have identified several growth factors and transcription factors with pivotal roles in the cardiogenic lineage commitment. Importantly, several of these factors contribute to multiple facets of cardiac development beyond just the early stages, such as for example chamber formation and valvulogenesis as reported for Nxk2.5 and Gata4 ([11][12][13]), respectively. Furthermore, the functional roles of these early cardiogenic lineage transcription factors are also associated to adult cardiac structural pathologies, such as dilated cardiomyopathy [221][222][223][224][225][226][227] or bicuspid aortic valves [228][229] and electrophysiological pathologies such as atrial fibrillation [230][231][232][233]. Manipulation of these core transcription factors have provided new tools to convert differentiated cells such as fibroblasts into cardiomyocytes [91], opening new therapeutic opportunities to heal the damaged heart. Importantly, a novel layer of gene regulation is emerging with the identification and functional characterization of non-coding RNAs that are important for cardiomyogenic commitment [234][235][236], broadening thus the therapeutic tools to provide novel approaches for cardiac repair.
The embryonic heart is the first organ to display morphological left–right asymmetry. Impairment of sidedness leads to severe body plan abnormalities, including herein cardiac defects. Our knowledge of the molecular and cellular bases of left-right symmetry break has considerably increased over the last decade, providing novel links between early molecular events and impaired sidedness . Such discoveries have served to unravel the genetic bases of heterotaxia and thus as guiding tools for genetic screening and counseling in these human conditions. The discovery of non-coding RNAs involved in early cardiac looping events also exemplify that the final picture of left–right signaling is still incomplete and it would anticipate novel discoveries in the front in the near future.
As cardiac looping takes place, the heart is progressively externally covered by the embryonic epicardium. Importantly, such embryonic epicardium will migrate and deepen into the embryonic myocardium leading to the formation of the cardiac fibroskeleton and part of the coronary vasculature . Failure on the formation of the embryonic epicardium or its progenitor cells in the proepicardium is incompatible with life since the ventricular walls are thinner and the coronary vasculature failed to properly form . Curiously, neither the proepicardium or the epicardium leads to significant contribution to the embryonic myocardium in vivo, but if isolated and cultured in vitro, they can do so . Such properties can be used to unlock the cardiomyogenic potential of the epicardium and thus to sever as a source of myocardial differentiation to heal the heart, as previously reported . Unlocking these events with non-coding RNAs have been recently reported , opening new strategies to heal the damaged heart.
The formation of the cardiac chambers and the cardiac conduction system is an intricated developmental process that is initiated right after left–right symmetry break and thus cardiac looping. Subsequently, the heart displays a complex septation process that provide morphogenetic cues to form a four-chambered organ with distinct inlet and outlet connections. A multitude of different growth factors such as Bmp, Fgf and Wnt family members, and of transcription factors participate on this cardiac developmental orchestra [44][45][46][47][48][49][50][51][52][53][54][55][56][57][58][59][60], and similarly as in the earlier events of heart formation, non–coding RNAs, including herein microRNAs and lncRNAs also participate [61][234][235][236], although our understanding of their functional role is still scarce. Deciphering the molecular cascades involved in these developmental processes have provided candidate genes to test and identify genetic culprits of congenital cardiac anomalies in humans, and thus to design strategies for genetic screening and counseling.
References
- Inman, K.E.; Downs, K.M. Localization of Brachyury (T) in embryonic and extraembryonic tissues during mouse gastrulation. Gene Expr. Patterns 2006, 6, 783–793.
- Saga, Y.; Miyagawa-Tomita, S.; Takagi, A.; Kitajima, S.; I Miyazaki, J.; Inoue, T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development 1999, 126, 3437–3447.
- Saga, Y.; Kitajima, S.; Miyagawa-Tomita, S. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc. Med. 2000, 10, 345–352.
- Kitajima, S.; Takagi, A.; Inoue, T.; Saga, Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development 2000, 127, 3215–3226.
- Bondue, A.; Lapouge, G.; Paulissen, C.; Semeraro, C.; Iacovino, M.; Kyba, M.; Blanpain, C. Mesp1 Acts as a Master Regulator of Multipotent Cardiovascular Progenitor Specification. Cell Stem Cell 2008, 3, 69–84.
- Bondue, A.; Blanpain, C. Mesp1: A key regulator of cardiovascular lineage commitment. Circ. Res. 2010, 107, 1414–1427.
- Chan, S.S.-K.; Shi, X.; Toyama, A.; Arpke, R.W.; Dandapat, A.; Iacovino, M.; Kang, J.-J.; Le, G.; Hagen, H.R.; Garry, D.J.; et al. Mesp1 Patterns Mesoderm into Cardiac, Hematopoietic, or Skeletal Myogenic Progenitors in a Context-Dependent Manner. Cell Stem Cell 2013, 12, 587–601.
- Lescroart, F.; Chabab, S.; Lin, X.; Rulands, S.; Paulissen, C.; Rodolosse, A.; Auer, H.; Achouri, Y.; Dubois, C.; Bondue, A.; et al. Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat. Cell Biol. 2014, 16, 829–840.
- Chiapparo, G.; Lin, X.; Lescroart, F.; Chabab, S.; Paulissen, C.; Pitisci, L.; Bondue, A.; Blanpain, C. Mesp1 controls the speed, polarity, and directionality of cardiovascular progenitor migration. J. Cell Biol. 2016, 213, 463–477.
- Laverriere, A.C.; MacNeill, C.; Mueller, C.; Poelmann, R.E.; Burch, J.B.; Evans, T. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J. Biol. Chem. 1994, 269, 23177–23184.
- Charron, F.; Nemer, M. GATA transcription factors and cardiac development. Semin. Cell Dev. Biol. 1999, 10, 85–91.
- Burch, J.B. Regulation of GATA gene expression during vertebrate development. Semin. Cell Dev. Biol. 2005, 16, 71–81.
- Brewer, A.; Pizzey, J. GATA factors in vertebrate heart development and disease. Expert Rev. Mol. Med. 2006, 8, 1–20.
- Jiang, Y.; Evans, T. The Xenopus GATA-4/5/6 genes are associated with cardiac specification and can regulate cardiac-specific transcription during embryogenesis. Dev. Biol. 1996, 174, 258–270.
- Lee, K.H.; Xu, Q.; Breitbart, R.E. A new tinman-related gene, nkx2.7, anticipate the expression of nkx2.5 and nkx2.3 in zebrafish heart and pharyngeal endoderm. Dev. Biol. 1996, 180, 722–731.
- Holtzinger, A.; Evans, T. Gata5 and Gata6 are functionally redundant in zebrafish for specification of cardiomyocytes. Dev. Biol. 2007, 312, 613–622.
- Sam, J.; Mercer, E.J.; Torregroza, I.; Banks, K.M.; Evans, T. Specificity, redundancy and dosage thresholds among gata4/5/6 genes during zebrafish cardiogenesis. Biol. Open 2020, 9, bio053611.
- Reecy, J.; Yamada, M.; Cummings, K.; Sosic, D.; Chen, C.-Y.; Eichele, G.; Olson, E.N.; Schwartz, R.J. Chicken Nkx-2.8: A Novel Homeobox Gene Expressed in Early Heart Progenitor Cells and Pharyngeal Pouch-2 and -3 Endoderm. Dev. Biol. 1997, 188, 295–311.
- Boettger, T.; Stein, S.; Kessel, M. The chicken NKX2.8 homeobox gene: A novel member of the NK-2 gene family. Dev. Genes Evol. 1997, 207, 65–70.
- Brand, T.; Andrée, B.; Schneider, A.; Buchberger, A.; Arnold, H.-H. Chicken NKx2-8, a novel homeobox gene expressed during early heart and foregut development. Mech. Dev. 1997, 64, 53–59.
- Jiang, Y.; Tarzami, S.; Burch, J.B.; Evans, T. Common role for each of the cGATA-4/5/6 genes in the regulation of cardiac morphogenesis. Dev. Genet. 1998, 22, 263–277.
- Ban, Q.; Liu, X.; Hui, W.; Chen, D.; Zhao, Z.; Jia, B. Comparative Analysis of Nkx2-5/GATA4/TBX5 Expression in Chicken, Quail and Chicken-quail Hybrids during the Early Stage of Cardiac Development in Embryos. Asian-Australas. J. Anim. Sci. 2013, 26, 476–482.
- Heikinheimo, M.; Scandrett, J.M.; Wilson, D.B. Localization of transcription factor GATA-4 to regions of the mouse embryo involved in cardiac development. Dev. Biol. 1994, 164, 361–373.
- Harvey, R.P. NK-2Homeobox Genes and Heart Development. Dev. Biol. 1996, 178, 203–216.
- Morrisey, E.E.; Ip, H.S.; Lu, M.M.; Parmacek, M.S. GATA-6: A Zinc Finger Transcription Factor That Is Expressed in Multiple Cell Lineages Derived from Lateral Mesoderm. Dev. Biol. 1996, 177, 309–322.
- Morrisey, E.E.; Ip, H.S.; Tang, Z.; Lu, M.M.; Parmacek, M.S. GATA-5: A Transcriptional Activator Expressed in a Novel Temporally and Spatially-Restricted Pattern during Embryonic Development. Dev. Biol. 1997, 183, 21–36.
- Morrisey, E.E.; Ip, H.S.; Tang, Z.; Parmacek, M.S. GATA-4 activates transcription via two novel domains that are conserved within the GATA-4/5/6 subfamily. J. Biol. Chem. 1997, 272, 8515–8524.
- Von Both, I.; Silvestri, C.; Erdemir, T.; Lickert, H.; Walls, J.R.; Henkelman, R.M.; Rossant, J.; Harvey, R.P.; Attisano, L.; Wrana, J.L. Foxh1 is essential for development of the anterior heart field. Dev. Cell 2004, 7, 331–345.
- Tanaka, M.; Yamasaki, N.; Izumo, S. Phenotypic Characterization of the Murine Nkx2.6 Homeobox Gene by Gene Targeting. Mol. Cell. Biol. 2000, 20, 2874–2879.
- Caprioli, A.; Koyano-Nakagawa, N.; Iacovino, M.; Shi, X.; Ferdous, A.; Harvey, R.P.; Olson, E.N.; Kyba, M.; Garry, D.J. Nkx2-5 Represses Gata1 Gene Expression and Modulates the Cellular Fate of Cardiac Progenitors During Embryogenesis. Circulation 2011, 123, 1633–1641.
- Kasahara, H.; Bartunkova, S.; Schinke, M.; Tanaka, M.; Izumo, S. Cardiac and Extracardiac Expression of Csx/Nkx2.5 Homeodomain Protein. Circ. Res. 1998, 82, 936–946.
- Tanaka, M.; Chen, Z.; Bartunkova, S.; Yamasaki, N.; Izumo, S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development 1999, 126, 1269–1280.
- Jamali, M.; Rogerson, P.J.; Wilton, S.; Skerjanc, I.S. Nkx2–5 Activity Is Essential for Cardiomyogenesis. J. Biol. Chem. 2001, 276, 42252–42258.
- Harvey, R.P.; Lai, D.; Elliott, D.A.; Biben, C.; Solloway, M.; Prall, O.; Stennard, F.; Schindeler, A.; Groves, N.; Lavulo, L.; et al. Homeodomain Factor Nkx2-5 in Heart Development and Disease. Cold Spring Harb. Symp. Quant. Biol. 2002, 67, 107–114.
- Lin, Q.; Schwarz, J.; Bucana, C.; Olson, E.N. Control of Mouse Cardiac Morphogenesis and Myogenesis by Transcription Factor MEF2C. Science 1997, 276, 1404–1407.
- Bi, W.; Drake, C.J.; Schwarz, J.J. The Transcription Factor MEF2C-Null Mouse Exhibits Complex Vascular Malformations and Reduced Cardiac Expression of Angiopoietin 1 and VEGF. Dev. Biol. 1999, 211, 255–267.
- Karamboulas, C.; Dakubo, G.D.; Liu, J.; De Repentigny, Y.; Yutzey, K.; Wallace, V.A.; Kothary, R.; Skerjanc, I.S. Disruption of MEF2 activity in cardiomyoblasts inhibits cardiomyogenesis. J. Cell. Sci. 2006, 119 Pt 20, 4315–4321.
- Materna, S.C.; Sinha, T.; Barnes, R.M.; Van Bueren, K.L.; Black, B.L. Cardiovascular development and survival require Mef2c function in the myocardial but not the endothelial lineage. Dev. Biol. 2019, 445, 170–177.
- Gajewski, K.; Fossett, N.; Molkentin, J.D.; A Schulz, R. The zinc finger proteins Pannier and GATA4 function as cardiogenic factors in Drosophila. Development 1999, 126, 5679–5688.
- Pu, W.T.; Ishiwata, T.; Juraszek, A.L.; Ma, Q.; Izumo, S. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev. Biol. 2004, 275, 235–244.
- Zhao, R.; Watt, A.J.; Battle, M.A.; Li, J.; Bondow, B.J.; Duncan, S.A. Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Dev. Biol. 2008, 317, 614–619.
- Haworth, K.E.; Kotecha, S.; Mohun, T.J.; Latinkić, B.V. GATA4 and GATA5 are essential for heart and liver development in Xenopus embryos. BMC Dev. Biol. 2008, 8, 74.
- Martin, J.; Afouda, B.A.; Hoppler, S. Wnt/beta-catenin signalling regulates cardiomyogenesis via GATA transcription factors. J. Anat. 2010, 216, 92–107.
- Clement, J.H.; Fettes, P.; Knöchel, S.; Lef, J.; Knöchel, W. Bone morphogenetic protein 2 in the early development of Xenopus laevis. Mech. Dev. 1995, 52, 357–370.
- Ladd, A.N.; Yatskievych, T.A.; Antin, P.B. Regulation of avian cardiac myogenesis by activin/TGFbeta and bone morphogenetic proteins. Dev. Biol. 1998, 204, 407–419.
- Monzen, K.; Shiojima, I.; Hiroi, Y.; Kudoh, S.; Oka, T.; Takimoto, E.; Hayashi, D.; Hosoda, T.; Habara-Ohkubo, A.; Nakaoka, T.; et al. Bone Morphogenetic Proteins Induce Cardiomyocyte Differentiation through the Mitogen-Activated Protein Kinase Kinase Kinase TAK1 and Cardiac Transcription Factors Csx/Nkx-2.5 and GATA-4. Mol. Cell. Biol. 1999, 19, 7096–7105.
- Schlange, T.; Andrée, B.; Arnold, H.-H.; Brand, T. BMP2 is required for early heart development during a distinct time period. Mech. Dev. 2000, 91, 259–270.
- Christiaen, L.A.; Stolfi, A.; Levine, M. BMP signaling coordinates gene expression and cell migration during precardiac mesoderm development. Dev. Biol. 2010, 340, 179–187.
- Gavrilov, S.; Lacy, E. Genetic dissection of ventral folding morphogenesis in mouse: Embryonic visceral endoderm-supplied BMP2 positions head and heart. Curr. Opin. Genet. Dev. 2013, 23, 461–469.
- Reifers, F.; Walsh, E.C.; Léger, S.; Stainier, D.Y.; Brand, M. Induction and differentiation of the zebrafish heart requires fibroblast growth factor 8 (fgf8/acerebellar). Development 2000, 127, 225–235.
- Lopez-Sanchez, C.; Climent, V.; Schoenwolf, G.C.; Alvarez, I.S.; Garcia-Martinez, V. Induction of cardiogenesis by Hensen’s node and fibroblast growth factors. Cell Tissue Res. 2002, 309, 237–249.
- Alsan, B.H.; Schultheiss, T.M. Regulation of avian cardiogenesis by Fgf8 signaling. Development 2002, 129, 1935–1943.
- Ilagan, R.; Abu-Issa, R.; Brown, D.; Yang, Y.-P.; Jiao, K.; Schwartz, R.J.; Klingensmith, J.; Meyers, E.N. Fgf8 is required for anterior heart field development. Development 2006, 133, 2435–2445.
- Marvin, M.J.; Di Rocco, G.; Gardiner, A.; Bush, S.M.; Lassar, A.B. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 2001, 15, 316–327.
- Nakamura, T.; Sano, M.; Songyang, Z.; Schneider, M.D. A Wnt–and beta-catenin-dependent pathway for mammalian cardiac myogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 5834–5839.
- Eisenberg, L.M.; Eisenberg, C.A. Evaluating the role of Wnt signal transduction in promoting the development of the heart. Sci. World J. 2007, 7, 161–176.
- Foley, A.C.; Mercola, M. Heart induction by Wnt antagonists depends on the homeodomain transcription factor Hex. Genes Dev. 2005, 19, 387–396.
- Klaus, A.; Saga, Y.; Taketo, M.M.; Tzahor, E.; Birchmeier, W. Distinct roles of Wnt/beta-catenin and Bmp signaling during early cardiogenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 18531–18536.
- Liu, Z.; Li, T.; Liu, Y.; Jia, Z.; Li, Y.; Zhang, C.; Chen, P.; Ma, K.; Affara, N.; Zhou, C. WNT signaling promotes Nkx2.5 expression and early cardiomyogenesis via downregulation of Hdac1. Biochim. Biophys. Acta (BBA)–Bioenerg. 2009, 1793, 300–311.
- Jain, R.; Li, D.; Gupta, M.; Manderfield, L.J.; Ifkovits, J.L.; Wang, Q.; Liu, F.; Liu, Y.; Poleshko, A.; Padmanabhan, A.; et al. Integration of Bmp and Wnt signaling by Hopx specifies commitment of cardiomyoblasts. Science 2015, 348, aaa6071.
- Lopez-Sanchez, C.; Franco, D.; Bonet, F.; Garcia-Lopez, V.; Aranega, A.; Garcia-Martinez, V. Negative Fgf8-Bmp2 feed-back is regulated by miR-130 during early cardiac specification. Dev. Biol. 2015, 406, 63–73.
- Christoffels, V.M.; Habets, P.E.; Franco, D.; Campione, M.; de Jong, F.; Lamers, W.H.; Bao, Z.Z.; Palmer, S.; Biben, C.; Harvey, R.P.; et al. Chamber formation and morphogenesis in the developing mammalian heart. Dev. Biol. 2000, 223, 266–278.
- Meilhac, S.M.; Kelly, R.G.; Rocancourt, D.; Eloy-Trinquet, S.; Nicolas, J.-F.; Buckingham, M.E. A retrospective clonal analysis of the myocardium reveals two phases of clonal growth in the developing mouse heart. Development 2003, 130, 3877–3889.
- Meilhac, S.M.; Esner, M.; Kelly, R.G.; Nicolas, J.-F.; Buckingham, M.E. The Clonal Origin of Myocardial Cells in Different Regions of the Embryonic Mouse Heart. Dev. Cell 2004, 6, 685–698.
- Buckingham, M.E.; Meilhac, S.M.; Zaffran, S. Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 2005, 6, 826–835.
- Meilhac, S.M.; Lescroart, F.; Blanpain, C.; Buckingham, M.E. Cardiac cell lineages that form the heart. Cold Spring Harb. Perspect. Med. 2014, 4, a013888.
- Meilhac, S.M.; Buckingham, M. The deployment of cell lineages that form the mammalian heart. Nat. Rev. Cardiol. 2018, 15, 705–724.
- Zhang, L.; Nomura-Kitabayashi, A.; Sultana, N.; Cai, W.; Cai, X.; Moon, A.M.; Cai, C.-L. Mesodermal Nkx2.5 is necessary and sufficient for early second heart field development. Dev. Biol. 2014, 390, 68–79.
- Cai, C.-L.; Liang, X.; Shi, Y.; Chu, P.-H.; Pfaff, S.L.; Chen, J.; Evans, S.M. Isl1 Identifies a Cardiac Progenitor Population that Proliferates Prior to Differentiation and Contributes a Majority of Cells to the Heart. Dev. Cell 2003, 5, 877–889.
- Dyer, L.A.; Makadia, F.A.; Scott, A.; Pegram, K.; Hutson, M.R.; Kirby, M.L. BMP signaling modulates hedgehog-induced secondary heart field proliferation. Dev. Biol. 2010, 348, 167–176.
- Hinits, Y.; Pan, L.; Walker, C.; Dowd, J.; Moens, C.B.; Hughes, S.M. Zebrafish Mef2ca and Mef2cb are essential for both first and second heart field cardiomyocyte differentiation. Dev. Biol. 2012, 369, 199–210.
- Van Oort, R.J.; van Rooij, E.; Bourajjaj, M.; Schimmel, J.; Jansen, M.A.; van der Nagel, R.; Doevendans, P.A.; Schneider, M.D.; van Echteld, C.J.; De Windt, L.J. MEF2 activates a genetic program promoting chamber dilation and contractile dysfunction in calcineurin-induced heart failure. Circulation 2006, 114, 298–308.
- Oka, T.; Maillet, M.; Watt, A.J.; Schwartz, R.J.; Aronow, B.J.; Duncan, S.A.; Molkentin, J.D. Cardiac-Specific Deletion of Gata4 Reveals Its Requirement for Hypertrophy, Compensation, and Myocyte Viability. Circ. Res. 2006, 98, 837–845.
- Muñoz, J.P.; Collao, A.; Chiong, M.; Maldonado, C.; Adasme, T.; Carrasco, L.; Ocaranza, P.; Bravo-Sagua, R.; González, L.; Díaz-Araya, G.; et al. The transcription factor MEF2C mediates cardiomyocyte hypertrophy induced by IGF-1 signaling. Biochem. Biophys. Res. Commun. 2009, 388, 155–160.
- Liang, Q.; De Windt, L.J.; Witt, S.A.; Kimball, T.R.; Markham, B.E.; Molkentin, J.D. The Transcription Factors GATA4 and GATA6 Regulate Cardiomyocyte Hypertrophy in Vitro and in Vivo. J. Biol. Chem. 2001, 276, 30245–30253.
- Kontaraki, J.E.; Parthenakis, F.I.; Patrianakos, A.; Karalis, I.K.; Vardas, P.E. Altered expression of early cardiac marker genes in circulating cells of patients with hypertrophic cardiomyopathy. Cardiovasc. Pathol. 2007, 16, 329–335.
- Van Berlo, J.H.; Elrod, J.W.; van den Hoogenhof, M.M.; York, A.J.; Aronow, B.J.; Duncan, S.A.; Molkentin, J.D. The transcription factor GATA-6 regulates pathological cardiac hypertrophy. Circ Res. 2010, 107, 1032–1040.
- Coppola, A.; Romito, A.; Borel, C.; Gehrig, C.; Gagnebin, M.; Falconnet, E.; Izzo, A.; Altucci, L.; Banfi, S.; Antonarakis, S.E.; et al. Cardiomyogenesis is controlled by the miR-99a/let-7c cluster and epigenetic modifications. Stem Cell Res. 2014, 12, 323–337.
- Ménard, C.; Grey, C.; Méry, A.; Zeineddine, D.; Aimond, F.; Pucéat, M. Cardiac specification of embryonic stem cells. J. Cell. Biochem. 2004, 93, 681–687.
- Glass, C.; Singla, R.; Arora, A.; Singla, D.K. Mouse Embryonic Stem Cell-Derived Cardiac Myocytes in a Cell Culture Dish. Methods Mol. Biol. 2015, 1299, 145–152.
- Kawai, T.; Takahashi, T.; Esaki, M.; Ushikoshi, H.; Nagano, S.; Fujiwara, H.; Kosai, K. Efficient cardiomyogenic differentiation of embryonic stem cell by fibroblast growth factor 2 and bone morphogenetic protein 2. Circ. J. 2004, 68, 691–702.
- Zimmermann, W.-H. Embryonic and embryonic-like stem cells in heart muscle engineering. J. Mol. Cell. Cardiol. 2011, 50, 320–326.
- Kinney, M.A.; Sargent, C.Y.; McDevitt, T.C. Temporal Modulation of β-Catenin Signaling by Multicellular Aggregation Kinetics Impacts Embryonic Stem Cell Cardiomyogenesis. Stem Cells Dev. 2013, 22, 2665–2677.
- Maltsev, V.A.; Rohwedel, J.; Hescheler, J.; Wobus, A.M. Embryonic stem cells differentiate in vitro into cardiomyocytes representing sinusnodal, atrial and ventricular cell types. Mech. Dev. 1993, 44, 41–50.
- Paige, S.L.; Plonowska, K.; Xu, A.; Wu, S.M. Molecular regulation of cardiomyocyte differentiation. Circ. Res. 2015, 116, 341–353.
- Sluijter, J.P.; Van Mil, A.; Van Vliet, P.; Metz, C.H.; Liu, J.; Doevendans, P.A.; Goumans, M.-J. MicroRNA-1 and -499 Regulate Differentiation and Proliferation in Human-Derived Cardiomyocyte Progenitor Cells. Arter. Thromb. Vasc. Biol. 2010, 30, 859–868.
- Turbendian, H.K.; Gordillo, M.; Tsai, S.-Y.; Lu, J.; Kang, G.; Liu, T.-C.; Tang, A.; Liu, S.; Fishman, G.I.; Evans, T. GATA factors efficiently direct cardiac fate from embryonic stem cells. Development 2013, 140, 1639–1644.
- Zhang, Z.; Zhang, A.D.; Kim, L.J.; Nam, Y.-J. Ensuring expression of four core cardiogenic transcription factors enhances cardiac reprogramming. Sci. Rep. 2019, 9, 1–10.
- Schweizer, P.A.; Darche, F.F.; Ullrich, N.D.; Geschwill, P.; Greber, B.; Rivinius, R.; Seyler, C.; Müller-Decker, K.; Draguhn, A.; Utikal, J.; et al. Subtype-specific differentiation of cardiac pacemaker cell clusters from human induced pluripotent stem cells. Stem Cell Res. Ther. 2017, 8, 1–15.
- Kempf, H.; Zweigerdt, R. Scalable Cardiac Differentiation of Pluripotent Stem Cells Using Specific Growth Factors and Small Molecules. Adv. Biochem. Eng. Biotechnol. 2017, 163, 39–69.
- Hartung, S.; Schwanke, K.; Haase, A.; David, R.; Franz, W.-M.; Martin, U.; Zweigerdt, R. Directing Cardiomyogenic Differentiation of Human Pluripotent Stem Cells by Plasmid-Based Transient Overexpression of Cardiac Transcription Factors. Stem Cells Dev. 2013, 22, 1112–1125.
- Ieda, M.; Fu, J.-D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct Reprogramming of Fibroblasts into Functional Cardiomyocytes by Defined Factors. Cell 2010, 142, 375–386.
- Zhang, Z.; Li, H.; Ma, Z.; Feng, J.; Gao, P.; Dong, H.; Zhang, Z. Efficient cardiomyogenic differentiation of bone marrow mesenchymal stromal cells by combination of Wnt11 and bone morphogenetic protein 2. Exp. Biol. Med. 2012, 237, 768–776.
- Kuo, C.T.; E Morrisey, E.; Anandappa, R.; Sigrist, K.; Lu, M.M.; Parmacek, M.S.; Soudais, C.; Leiden, J.M. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997, 11, 1048–1060.
- Molkentin, J.D.; Lin, Q.; Duncan, S.A.; Olson, E.N. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997, 11, 1061–1072.
- Biben, C.; Harvey, R.P. Homeodomain factor Nkx2-5 controls left/right asymmetric expression of bHLH gene eHand during murine heart development. Genes Dev. 1997, 11, 1357–1369.
- Durocher, D.; Charron, F.; Warren, R.; Schwartz, R.J.; Nemer, M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997, 16, 5687–5696.
- Skerjanc, I.S.; Petropoulos, H.; Ridgeway, A.G.; Wilton, S. Myocyte enhancer factor 2C and Nkx2-5 up-regulate each other’s expression and initiate cardiomyogenesis in P19 cells. J. Biol. Chem. 1998, 273, 34904–34910.
- Shiojima, I.; Komuro, I.; Oka, T.; Hiroi, Y.; Mizuno, T.; Takimoto, E.; Monzen, K.; Aikawa, R.; Akazawa, H.; Yamazaki, T.; et al. Context-dependent Transcriptional Cooperation Mediated by Cardiac Transcription Factors Csx/Nkx-2.5 and GATA-4. J. Biol. Chem. 1999, 274, 8231–8239.
- Vincentz, J.W.; Barnes, R.M.; Firulli, B.A.; Conway, S.J.; Firulli, A.B. Cooperative interaction of Nkx2.5 and Mef2c transcription factors during heart development. Dev. Dyn. 2008, 237, 3809–3819.
- Ghosh, T.K.; Song, F.F.; Packham, E.A.; Buxton, S.; Robinson, T.E.; Ronksley, J.; Self, T.; Bonser, A.J.; Brook, J.D. Physical Interaction between TBX5 and MEF2C Is Required for Early Heart Development. Mol. Cell. Biol. 2009, 29, 2205–2218.
- Hiroi, Y.; Kudoh, S.; Monzen, K.; Ikeda, Y.; Yazaki, Y.; Nagai, R.; Komuro, I. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat. Genet. 2001, 28, 276–280.
- Maitra, M.; Schluterman, M.K.; Nichols, H.A.; Richardson, J.A.; Lo, C.W.; Srivastava, D.; Garg, V. Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev. Biol. 2009, 326, 368–377.
- Pradhan, L.; Gopal, S.; Li, S.; Ashur, S.; Suryanarayanan, S.; Kasahara, H.; Nam, H.-J. Intermolecular Interactions of Cardiac Transcription Factors NKX2.5 and TBX5. Biochemistry 2016, 55, 1702–1710.
- Singh, M.K.; Li, Y.; Li, S.; Cobb, R.M.; Zhou, D.; Lu, M.M.; Epstein, J.A.; Morrisey, E.E.; Gruber, P.J. Gata4 and Gata5 Cooperatively Regulate Cardiac Myocyte Proliferation in Mice. J. Biol. Chem. 2010, 285, 1765–1772.
- Charron, F.; Paradis, P.; Bronchain, O.; Nemer, G.; Nemer, M. Cooperative Interaction between GATA-4 and GATA-6 Regulates Myocardial Gene Expression. Mol. Cell. Biol. 1999, 19, 4355–4365.
- Prall, O.W.; Menon, M.K.; Solloway, M.J.; Watanabe, Y.; Zaffran, S.; Bajolle, F.; Biben, C.; McBride, J.J.; Robertson, B.R.; Chaulet, H.; et al. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell 2007, 128, 947–959.
- Schlesinger, J.; Schueler, M.; Grunert, M.; Fischer, J.J.; Zhang, Q.; Krueger, T.; Lange, M.; Tönjes, M.; Dunkel, I.; Sperling, S. The Cardiac Transcription Network Modulated by Gata4, Mef2a, Nkx2.5, Srf, Histone Modifications, and MicroRNAs. PLoS Genet. 2011, 7, e1001313.
- Zhou, L.; Liu, Y.; Lu, L.; Lu, X.; Dixon, R.A. Cardiac Gene Activation Analysis in Mammalian Non-Myoblasic Cells by Nkx2-5, Tbx5, Gata4 and Myocd. PLoS ONE 2012, 7, e48028.
- Lu, J.R.; McKinsey, T.A.; Xu, H.; Wang, D.Z.; Richardson, J.A.; Olson, E.N. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol. Cell. Biol. 1999, 19, 4495–4502.
- Akazawa, H.; Kudoh, S.; Mochizuki, N.; Takekoshi, N.; Takano, H.; Nagai, T.; Komuro, I. A novel LIM protein Cal promotes cardiac differentiation by association with CSX/NKX2-5. J. Cell Biol. 2004, 164, 395–405.
- Voronova, A.; Al Madhoun, A.; Fischer, A.; Shelton, M.; Karamboulas, C.; Skerjanc, I.S. Gli2 and MEF2C activate each other’s expression and function synergistically during cardiomyogenesis in vitro. Nucleic Acids Res. 2012, 40, 4723–4724.
- Behrens, A.N.; Iacovino, M.; Lohr, J.L.; Ren, Y.; Zierold, C.; Harvey, R.P.; Kyba, M.; Garry, D.J.; Martin, C.M. Nkx2-5 Mediates Differential Cardiac Differentiation Through Interaction with Hoxa10. Stem Cells Dev. 2013, 22, 2211–2220.
- Clark, C.D.; Lee, K.-H. Second heart field-specific expression of Nkx2-5 requires promoter proximal interaction with Srf. Mech. Dev. 2020, 162, 103615.
- Alexandrovich, A.; Arno, M.; Patient, R.K.; Shah, A.M.; Pizzey, J.A.; Brewer, A.C. Wnt2 is a direct downstream target of GATA6 during early cardiogenesis. Mech. Dev. 2006, 123, 297–311.
- Behrens, A.N.; Ren, Y.; Ferdous, A.; Garry, D.J.; Martin, C.M. Nkx2-5 Regulates Tdgf1 (Cripto) Early During Cardiac Development. J. Clin. Exp. Cardiol. 2013, 1, 1–4.
- Cambier, L.; Plate, M.; Sucov, H.M.; Pashmforoush, M. Nkx2-5 regulates cardiac growth through modulation of Wnt signaling by R-spondin3. Development 2014, 141, 2959–2971.
- Dodou, E.; Verzi, M.P.; Anderson, J.P.; Xu, S.-M.; Black, B.L. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development 2004, 131, 3931–3942.
- Dorn, T.; Goedel, A.; Lam, J.T.; Haas, J.; Tian, Q.; Herrmann, F.; Bundschu, K.; Dobreva, G.; Schiemann, M.; Dirschinger, R.; et al. Direct Nkx2-5 Transcriptional Repression of Isl1 Controls Cardiomyocyte Subtype Identity. Stem Cells 2015, 33, 1113–1129.
- Anderson, D.J.; Kaplan, D.I.; Bell, K.M.; Koutsis, K.; Haynes, J.M.; Mills, R.J.; Phelan, D.G.; Qian, E.L.; Leitoguinho, A.R.; Arasaratnam, D.; et al. NKX2-5 regulates human cardiomyogenesis via a HEY2 dependent transcriptional network. Nat. Commun. 2018, 9, 1–13.
- Horton, A.J.; Brooker, J.; Streitfeld, W.S.; Flessa, M.E.; Pillai, B.; Simpson, R.; Clark, C.D.; Gooz, M.B.; Sutton, K.K.; Foley, A.C.; et al. Nkx2–5 Second Heart Field Target Gene Ccdc117 Regulates DNA Metabolism and Proliferation. Sci. Rep. 2019, 9, 1–12.
- Liu, Z.-P.; Nakagawa, O.; Nakagawa, M.; Yanagisawa, H.; Passier, R.; Richardson, J.A.; Srivastava, D.; Olson, E.N. CHAMP, A Novel Cardiac-Specific Helicase Regulated by MEF2C. Dev. Biol. 2001, 234, 497–509.
- Qian, L.; Wythe, J.D.; Liu, J.; Cartry, J.; Vogler, G.; Mohapatra, B.; Otway, R.T.; Huang, Y.; King, I.N.; Maillet, M.; et al. Tinman/Nkx2-5 acts via miR-1 and upstream of Cdc42 to regulate heart function across species. J. Cell Biol. 2011, 193, 1181–1196.
- Yu, B.; Gong, M.; Wang, Y.; Millard, R.W.; Pasha, Z.; Yang, Y.; Ashraf, M.; Xu, M. Cardiomyocyte Protection by GATA-4 Gene Engineered Mesenchymal Stem Cells Is Partially Mediated by Translocation of miR-221 in Microvesicles. PLoS ONE 2013, 8, e73304.
- Watt, A.J.; Battle, M.A.; Li, J.; Duncan, S.A. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc. Natl. Acad. Sci. USA 2004, 101, 12573–12578.
- Zhou, B.; Von Gise, A.; Ma, Q.; Rivera-Feliciano, J.; Pu, W.T. Nkx2-5- and Isl1-expressing cardiac progenitors contribute to proepicardium. Biochem. Biophys. Res. Commun. 2008, 375, 450–453.
- Zeisberg, E.M.; Ma, Q.; Juraszek, A.L.; Moses, K.; Schwartz, R.J.; Izumo, S.; Pu, W.T. Morphogenesis of the right ventricle requires myocardial expression of Gata4. J. Clin. Investig. 2005, 115, 1522–1531.
- Moskowitz, I.P.; Kim, J.B.; Moore, M.L.; Wolf, C.M.; Peterson, M.A.; Shendure, J.; Nóbrega, M.A.; Yokota, Y.; Berul, C.; Izumo, S.; et al. A Molecular Pathway Including Id2, Tbx5, and Nkx2-5 Required for Cardiac Conduction System Development. Cell 2007, 129, 1365–1376.
- Espinoza-Lewis, R.A.; Liu, H.; Sun, C.; Chen, C.; Jiao, K.; Chen, Y.-P. Ectopic expression of Nkx2.5 suppresses the formation of the sinoatrial node in mice. Dev. Biol. 2011, 356, 359–369.
- Jay, P.Y.; Harris, B.S.; Maguire, C.T.; Buerger, A.; Wakimoto, H.; Tanaka, M.; Kupershmidt, S.; Roden, D.M.; Schultheiss, T.M.; O’Brien, T.X.; et al. Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J. Clin. Investig. 2004, 113, 1130–1137.
- Meysen, S.; Marger, L.; Hewett, K.W.; Jarry-Guichard, T.; Agarkova, I.; Chauvin, J.P.; Perriard, J.C.; Izumo, S.; Gourdie, R.G.; Mangoni, M.E.; et al. Nkx2.5 cell-autonomous gene function is required for the postnatal formation of the peripheral ventricular conduction system. Dev. Biol. 2007, 303, 740–753.
- Harris, B.S.; Spruill, L.; Edmonson, A.M.; Rackley, M.S.; Benson, D.W.; O’Brien, T.X.; Gourdie, R.G. Differentiation of cardiac Purkinje fibers requires precise spatiotemporal regulation of Nkx2-5 expression. Dev. Dyn. 2005, 235, 38–49.
- Biben, C.; Weber, R.; Kesteven, S.; Stanley, E.; McDonald, L.; Elliott, D.A.; Barnett, L.; Köentgen, F.; Robb, L.; Feneley, M.; et al. Cardiac Septal and Valvular Dysmorphogenesis in Mice Heterozygous for Mutations in the Homeobox GeneNkx2-5. Circ. Res. 2000, 87, 888–895.
- Tanaka, M.; Berul, C.; Ishii, M.; Jay, P.; Wakimoto, H.; Douglas, P.; Yamasaki, N.; Kawamoto, T.; Gehrmann, J.; Maguire, C.; et al. A Mouse Model of Congenital Heart Disease: Cardiac Arrhythmias and Atrial Septal Defect Caused by Haploinsufficiency of the Cardiac Transcription Factor Csx/Nkx2.5. Cold Spring Harb. Symp. Quant. Biol. 2002, 67, 317–326.
- Terada, R.; Warren, S.; Lu, J.T.; Chien, K.R.; Wessels, A.; Kasahara, H. Ablation of Nkx2-5 at mid-embryonic stage results in premature lethality and cardiac malformation. Cardiovasc. Res. 2011, 91, 289–299.
- Phan, D.; Rasmussen, T.L.; Nakagawa, O.; McAnally, J.; Gottlieb, P.D.; Tucker, P.W.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. BOP, a regulator of right ventricular heart development, is a direct transcriptional target of MEF2C in the developing heart. Development 2005, 132, 2669–2678.
- Liberatore, C.M.; Searcy-Schrick, R.D.; Yutzey, K.E. Ventricular Expression of tbx5 Inhibits Normal Heart Chamber Development. Dev. Biol. 2000, 223, 169–180.
- Koshiba-Takeuchi, K.; Mori, A.D.; Kaynak, B.L.; Cebra-Thomas, J.; Sukonnik, T.; Georges, R.O.; Latham, S.; Beck, L.; Henkelman, R.M.; Black, B.L.; et al. Reptilian heart development and the molecular basis of cardiac chamber evolution. Nature 2009, 461, 95–98.
- Zhang, K.K.; Xiang, M.; Zhou, L.; Liu, J.; Curry, N.; Heine, S.D.; Garcia-Pavia, P.; Zhang, X.; Wang, Q.; Xie, L. Gene network and familial analyses uncover a gene network involving Tbx5/Osr1/Pcsk6 interaction in the second heart field for atrial septation. Hum. Mol. Genet. 2016, 25, 1140–1151.
- Nadeaua, M.; Georges, R.O.; Laforest, B.; Yamak, A.; Lefebvre, C.; Beauregard, J.; Paradis, P.; Bruneau, B.G.; Andelfinger, G.; Nemer, M. An endocardial pathway involving Tbx5, Gata4, and Nos3 required for atrial septum formation. Proc. Natl. Acad. Sci. USA 2010, 107, 19356–19361.
- Xie, L.; Hoffmann, A.D.; Burnicka-Turek, O.; Friedland-Little, J.M.; Zhang, K.; Moskowitz, I.P. Tbx5-Hedgehog Molecular Networks Are Essential in the Second Heart Field for Atrial Septation. Dev. Cell 2012, 23, 280–291.
- Misra, C.; Chang, S.-W.; Basu, M.; Huang, N.; Garg, V. Disruption of myocardial Gata4 and Tbx5 results in defects in cardiomyocyte proliferation and atrioventricular septation. Hum. Mol. Genet. 2014, 23, 5025–5035.
- Campione, M.; Steinbeisser, H.; Schweickert, A.; Deissler, K.; Van Bebber, F.; A Lowe, L.; Nowotschin, S.; Viebahn, C.; Haffter, P.; Kuehn, M.R.; et al. The homeobox gene Pitx2: Mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development 1999, 126, 1225–1234.
- Piedra, M.; Icardo, J.M.; Albajar, M.; Rodriguez-Rey, J.C.; A Ros, M. Pitx2 Participates in the Late Phase of the Pathway Controlling Left-Right Asymmetry. Cell 1998, 94, 319–324.
- Ryan, A.K.; Blumberg, B.; Rodriguez-Esteban, C.; Yonei-Tamura, S.; Tamura, K.; Tsukui, T.; De La Peña, J.; Sabbagh, W.; Greenwald, J.; Choe, S.; et al. Pitx2 determines left–right asymmetry of internal organs in vertebrates. Nat. Cell Biol. 1998, 394, 545–551.
- Logan, M.; Pagán-Westphal, S.M.; Smith, D.M.; Paganessi, L.; Tabin, C.J. The Transcription Factor Pitx2 Mediates Situs-Specific Morphogenesis in Response to Left-Right Asymmetric Signals. Cell 1998, 94, 307–317.
- Yoshioka, H.; Meno, C.; Koshiba, K.; Sugihara, M.; Itoh, H.; Ishimaru, Y.; Inoue, T.; Ohuchi, H.; Semina, E.V.; Murray, J.C.; et al. Pitx2, a Bicoid-Type Homeobox Gene, Is Involved in a Lefty-Signaling Pathway in Determination of Left-Right Asymmetry. Cell 1998, 94, 299–305.
- Bisgrove, B.W.; Essner, J.J.; Yost, H.J. Multiple pathways in the midline regulate concordant brain, heart and gut left-right asymmetry. Development 2000, 127, 3567–3579.
- Collins, M.M.; Maischein, H.-M.; Dufourcq, P.; Charpentier, M.; Blader, P.; Stainier, D.Y. Pitx2c orchestrates embryonic axis extension via mesendodermal cell migration. eLife 2018, 7, e34880.
- Dagle, J.; Sabel, J.L.; Littig, J.L.; Sutherland, L.B.; Kolker, S.J.; Weeks, D.L. Pitx2c attenuation results in cardiac defects and abnormalities of intestinal orientation in developing Xenopus laevis. Dev. Biol. 2003, 262, 268–281.
- Essner, J.J.; Branford, W.W.; Zhang, J.; Yost, H.J. Mesendoderm and left-right brain, heart and gut development are differentially regulated by pitx2 isoforms. Development 2000, 127, 1081–1093.
- Gage, P.J.; Suh, H.; Camper, S.A. Dosage requirement of Pitx2 for development of multiple organs. Development 1999, 126, 4643–4651.
- Yu, X.; Amand, T.R.S.; Wang, S.; Li, G.; Zhang, Y.; Hu, Y.P.; Nguyen, L.; Qiu, M.S.; Chen, Y.P. Differential expression and functional analysis of Pitx2 isoforms in regulation of heart looping in the chick. Development 2001, 128, 1005–1013.
- Lu, M.-F.; Pressman, C.L.; Dyer, R.; Johnson, R.L.; Martin, J.F. Function of Rieger syndrome gene in left–right asymmetry and craniofacial development. Nat. Cell Biol. 1999, 401, 276–278.
- Kitamura, K.; Miura, H.; Miyagawa-Tomita, S.; Yanazawa, M.; Katoh-Fukui, Y.; Suzuki, R.; Ohuchi, H.; Suehiro, A.; Motegi, Y.; Nakahara, Y.; et al. Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development 1999, 126, 5749–5758.
- Shiratori, H.; Yashiro, K.; Shen, M.M.; Hamada, H. Conserved regulation and role of Pitx2 in situs-specific morphogenesis of visceral organs. Development 2006, 133, 3015–3025.
- A Lowe, L.; Yamada, S.; Kuehn, M.R. Genetic dissection of nodal function in patterning the mouse embryo. Development 2001, 128, 1831–1843.
- St Amand, T.R.; Ra, J.; Zhang, Y.; Hu, Y.; Baber, S.I.; Qiu, M.; Chen, Y. Cloning and expression pattern of chicken Pitx2: A new component in the SHH signaling pathway controlling embryonic heart looping. Biochem. Biophys. Res. Commun. 1998, 247, 100–105.
- Campione, M.; A Ros, M.; Icardo, J.M.; Piedra, E.; Christoffels, V.M.; Schweickert, A.; Blum, M.; Franco, D.; Moorman, A.F. Pitx2 Expression Defines a Left Cardiac Lineage of Cells: Evidence for Atrial and Ventricular Molecular Isomerism in the iv/iv Mice. Dev. Biol. 2001, 231, 252–264.
- Shiratori, H.; Sakuma, R.; Watanabe, M.; Hashiguchi, H.; Mochida, K.; Sakai, Y.; Nishino, J.; Saijoh, Y.; Whitman, M.; Hamada, H. Two-step regulation of left-right asymmetric expression of Pitx2: Initiation by nodal signaling and maintenance by Nkx2. Mol. Cell. 2001, 7, 137–149.
- Campione, M.; Acosta, L.; Martinez, S.; Icardo, J.; Aranega, A.; Franco, D. Pitx2 and Cardiac Development: A Molecular Link between Left/Right Signaling and Congenital Heart Disease. Cold Spring Harb. Symp. Quant. Biol. 2002, 67, 89–95.
- Franco, D.; Campione, M. The role of Pitx2 during cardiac development. Linking left-right signaling and congenital heart diseases. Trends Cardiovasc. Med. 2003, 13, 157–163.
- Campione, M.; Franco, D. Current Perspectives in Cardiac Laterality. J. Cardiovasc. Dev. Dis. 2016, 3, 34.
- Franco, D.; Sedmera, D.; Lozano-Velasco, E. Multiple Roles of Pitx2 in Cardiac Development and Disease. J. Cardiovasc. Dev. Dis. 2017, 4, 16.
- Ai, D.; Liu, W.; Ma, L.; Dong, F.; Lu, M.-F.; Wang, D.; Verzi, M.P.; Cai, C.; Gage, P.J.; Evans, S.; et al. Pitx2 regulates cardiac left–right asymmetry by patterning second cardiac lineage-derived myocardium. Dev. Biol. 2006, 296, 437–449.
- Franco, D.; Campione, M.; Kelly, R.G.; Zammit, P.S.; Buckingham, M.; Lamers, W.H.; Moorman, A.F.M. Multiple transcriptional domains, with distinct left and right components, in the atrial chambers of the developing heart. Circ. Res. 2000, 87, 984–991.
- Mommersteeg, M.T.M.; Hoogaars, W.M.H.; Prall, O.W.J.; Vries, C.D.G.-D.; Wiese, C.; Clout, D.E.W.; Papaioannou, V.E.; Brown, N.A.; Harvey, R.P.; Moorman, A.F.M.; et al. Molecular Pathway for the Localized Formation of the Sinoatrial Node. Circ. Res. 2007, 100, 354–362.
- Ocaña, O.H.; Coskun, H.; Minguillón, C.; Murawala, P.; Tanaka, E.M.; Galcerán, J.; Muñoz-Chápuli, R.; Nieto, M.A. A right-handed signalling pathway drives heart looping in vertebrates. Nature 2017, 549, 86–90.
- Zeng, B.; Ren, X.-F.; Cao, F.; Zhou, X.-Y.; Zhang, J. Developmental patterns and characteristics of epicardial cell markers Tbx18 and Wt1 in murine embryonic heart. J. Biomed. Sci. 2011, 18, 67.
- Moore, A.W.; McInnes, L.; Kreidberg, J.; Hastie, N.D.; Schedl, A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development 1999, 126, 1845–1857.
- Acharya, A.; Baek, S.T.; Huang, G.; Eskiocak, B.; Goetsch, S.; Sung, C.Y.; Banfi, S.; Sauer, M.F.; Olsen, G.S.; Duffield, J.S.; et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development 2012, 139, 2139–2149.
- Ishii, Y.; Langberg, J.D.; Hurtado, R.; Lee, S.; Mikawa, T. Induction of proepicardial marker gene expression by the liver bud. Development 2007, 134, 3627–3637.
- Singh, R.; Hoogaars, W.M.; Barnett, P.; Grieskamp, T.; Rana, M.S.; Buermans, H.; Farin, H.F.; Petry, M.; Heallen, T.; Martin, J.F.; et al. Tbx2 and Tbx3 induce atrioventricular myocardial development and endocardial cushion formation. Cell Mol. Life Sci. 2012, 69, 1377–1389.
- Christoffels, V.M.; Hoogaars, W.M.; Tessari, A.; Clout, D.E.; Moorman, A.F.M.; Campione, M. T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Dev. Dyn. 2004, 229, 763–770.
- Sedletcaia, A.; Evans, T. Heart chamber size in zebrafish is regulated redundantly by duplicated tbx2 genes. Dev. Dyn. 2011, 240, 1548–1557.
- Chi, N.C.; Shaw, R.M.; De Val, S.; Kang, G.; Jan, L.Y.; Black, B.L.; Stainier, D.Y. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 2008, 22, 734–739.
- Shirai, M.; Imanaka-Yoshida, K.; Schneider, M.D.; Schwartz, R.J.; Morisaki, T. T-box 2, a mediator of Bmp-Smad signaling, induced hyaluronan synthase 2 and Tgfbeta2 expression and endocardial cushion formation. Proc. Natl. Acad. Sci. USA 2009, 106, 18604–18609.
- Dupays, L.; Kotecha, S.; Angst, B.; Mohun, T.J. Tbx2 misexpression impairs deployment of second heart field derived progenitor cells to the arterial pole of the embryonic heart. Dev. Biol. 2009, 333, 121–131.
- Hoogaars, W.M.; Tessari, A.; Moorman, A.F.; de Boer, P.A.; Hagoort, J.; Soufan, A.T.; Campione, M.; Christoffels, V.M. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc. Res. 2004, 62, 489–499.
- Mesbah, K.; Harrelson, Z.; Théveniau-Ruissy, M.; Papaioannou, V.E.; Kelly, R.G. Tbx3 Is Required for Outflow Tract Development. Circ. Res. 2008, 103, 743–750.
- Mohan, R.A.; Mommersteeg, M.T.M.; Domínguez, J.N.; Choquet, C.; Wakker, V.; de Gier-de Vries, C.; Boink, G.J.J.; Boukens, B.J.; Miquerol, L.; Verkerk, A.O.; et al. Embryonic Tbx3+ cardiomyocytes form the mature cardiac conduction system by progressive fate restriction. Development 2018, 145, dev167361.
- Mohan, R.A.; Bosada, F.M.; Van Weerd, J.H.; Van Duijvenboden, K.; Wang, J.; Mommersteeg, M.T.; Hooijkaas, I.B.; Wakker, V.; Vries, C.D.G.-D.; Coronel, R.; et al. T-box transcription factor 3 governs a transcriptional program for the function of the mouse atrioventricular conduction system. Proc. Natl. Acad. Sci. USA 2020, 117, 18617–18626.
- Cai, X.; Nomura-Kitabayashi, A.; Cai, W.; Yan, J.; Christoffels, V.M.; Cai, C.-L. Myocardial Tbx20 regulates early atrioventricular canal formation and endocardial epithelial–mesenchymal transition via Bmp2. Dev. Biol. 2011, 360, 381–390.
- Singh, M.K.; Christoffels, V.M.; Dias, J.M.; Trowe, M.-O.; Petry, M.; Schuster-Gossler, K.; Bürger, A.; Ericson, J.; Kispert, A. Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development 2005, 132, 2697–2707.
- Singh, R.; Horsthuis, T.; Farin, H.F.; Grieskamp, T.; Norden, J.; Petry, M.; Wakker, V.; Moorman, A.F.M.; Christoffels, V.M.; Kispert, A. Tbx20 Interacts With Smads to Confine Tbx2 Expression to the Atrioventricular Canal. Circ. Res. 2009, 105, 442–452.
- Stennard, F.A.; Costa, M.W.; Lai, D.; Biben, C.; Furtado, M.B.; Solloway, M.J.; McCulley, D.J.; Leimena, C.; Preis, J.I.; Dunwoodie, S.L.; et al. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development 2005, 132, 2451–2462.
- Chakraborty, S.; Yutzey, K.E. Tbx20 regulation of cardiac cell proliferation and lineage specialization during embryonic and fetal development in vivo. Dev. Biol. 2012, 363, 234–246.
- Greulich, F.; Rudat, C.; Kispert, A. Mechanisms of T-box gene function in the developing heart. Cardiovasc. Res. 2011, 91, 212–222.
- Plageman, T.F.; Yutzey, K.E. T-box genes and heart development: Putting the “T” in heart. Dev. Dyn. 2005, 232, 11–20.
- Cai, C.-L.; Zhou, W.; Yang, L.; Bu, L.; Qyang, Y.; Zhang, X.; Li, X.; Rosenfeld, M.G.; Chen, J.; Evans, S.M. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development 2005, 132, 2475–2487.
- Christoffels, V.M.; Keijser, A.G.; Houweling, A.C.; Clout, D.E.; Moorman, A.F. Patterning the Embryonic Heart: Identification of Five Mouse Iroquois Homeobox Genes in the Developing Heart. Dev. Biol. 2000, 224, 263–274.
- Kim, K.-H.; Rosen, A.; Bruneau, B.G.; Hui, C.-C.; Backx, P.H. Iroquois Homeodomain Transcription Factors in Heart Development and Function. Circ. Res. 2012, 110, 1513–1524.
- Bruneau, B.G.; Bao, Z.-Z.; Tanaka, M.; Schott, J.-J.; Izumo, S.; Cepko, C.L.; Seidman, J.; Seidman, C.E. Cardiac Expression of the Ventricle-Specific Homeobox Gene Irx4 Is Modulated by Nkx2-5 and dHand. Dev. Biol. 2000, 217, 266–277.
- Kokubo, H.; Miyagawa-Tomita, S.; Nakazawa, M.; Saga, Y.; Johnson, R.L. Mouse hesr1 and hesr2 genes are redundantly required to mediate Notch signaling in the developing cardiovascular system. Dev. Biol. 2005, 278, 301–309.
- Kokubo, H.; Tomita-Miyagawa, S.; Hamada, Y.; Saga, Y. Hesr1 and Hesr2 regulate atrioventricular boundary formation in the developing heart through the repression of Tbx2. Development 2007, 134, 747–755.
- Rutenberg, J.B.; Fischer, A.; Jia, H.; Gessler, M.; Zhong, T.P.; Mercola, M. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development 2006, 133, 4381–4390.
- Koibuchi, N.; Chin, M.T. CHF1/Hey2 Plays a Pivotal Role in Left Ventricular Maturation Through Suppression of Ectopic Atrial Gene Expression. Circ. Res. 2007, 100, 850–855.
- Pereira, F.A.; Qiu, Y.; Zhou, G.; Tsai, M.-J.; Tsai, S.Y. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev. 1999, 13, 1037–1049.
- Srivastava, D.; Cserjesi, P.; Olson, E.N. A Subclass of bHLH Proteins Required for Cardiac Morphogenesis. Science 1995, 270, 1995–1999.
- Srivastava, D.; Thomas, T.; Lin, Q.; Kirby, M.L.; Brown, R.; Olson, E.N. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat. Genet. 1997, 16, 154–160.
- Togi, K.; Kawamoto, T.; Yamauchi, R.; Yoshida, Y.; Kita, T.; Tanaka, M. Role of Hand1/eHAND in the Dorso-Ventral Patterning and Interventricular Septum Formation in the Embryonic Heart. Mol. Cell. Biol. 2004, 24, 4627–4635.
- Yamagishi, H.; Yamagishi, C.; Nakagawa, O.; Harvey, R.P.; Olson, E.N.; Srivastava, D. The combinatorial activities of Nkx2.5 and dHAND are essential for cardiac ventricle formation. Dev. Biol. 2001, 239, 190–203.
- Bolte, C.; Zhang, Y.; Wang, I.-C.; Kalin, T.V.; Molkentin, J.D.; Kalinichenko, V.V. Expression of Foxm1 Transcription Factor in Cardiomyocytes Is Required for Myocardial Development. PLoS ONE 2011, 6, e22217.
- Chen, F.; Kook, H.; Milewski, R.; Gitler, A.D.; Lu, M.M.; Li, J.; Nazarian, R.; Schnepp, R.; Jen, K.; Biben, C.; et al. Hop Is an Unusual Homeobox Gene that Modulates Cardiac Development. Cell 2002, 110, 713–723.
- Lavallée, G.; Andelfinger, G.; Nadeau, M.; Lefebvre, C.; Nemer, G.; E Horb, M.; Nemer, M. The Kruppel-like transcription factor KLF13 is a novel regulator of heart development. EMBO J. 2006, 25, 5201–5213.
- Niu, Z.; Yu, W.; Zhang, S.X.; Barron, M.; Belaguli, N.S.; Schneider, M.D.; Parmacek, M.; Nordheim, A.; Schwartz, R.J. Conditional Mutagenesis of the Murine Serum Response Factor Gene Blocks Cardiogenesis and the Transcription of Downstream Gene Targets. J. Biol. Chem. 2005, 280, 32531–32538.
- Parlakian, A.; Tuil, D.; Hamard, G.; Tavernier, G.; Hentzen, D.; Concordet, J.-P.; Paulin, D.; Li, Z.; Daegelen, D. Targeted Inactivation of Serum Response Factor in the Developing Heart Results in Myocardial Defects and Embryonic Lethality. Mol. Cell. Biol. 2004, 24, 5281–5289.
- Espinoza-Lewis, R.A.; Yu, L.; He, F.; Liu, H.; Tang, R.; Shi, J.; Sun, X.; Martin, J.F.; Wang, D.; Yang, J.; et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev. Biol. 2009, 327, 376–385.
- Hu, W.; Xin, Y.; Zhao, Y.; Hu, J. Shox2: The Role in Differentiation and Development of Cardiac Conduction System. Tohoku J. Exp. Med. 2018, 244, 177–186.
- Ye, W.; Wang, J.; Song, Y.; Yu, D.; Sun, C.; Liu, C.; Chen, F.; Zhang, Y.; Wang, F.; Harvey, R.P.; et al. A common Shox2-Nkx2-5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Development 2015, 142, 2521–2532.
- Liu, H.; Espinoza-Lewis, R.A.; Chen, C.; Hu, X.; Zhang, Y.; Chen, Y.-P. The Role of Shox2 in SAN Development and Function. Pediatr. Cardiol. 2012, 33, 882–889.
- Liu, H.; Chen, C.-H.; Espinoza-Lewis, R.A.; Jiao, Z.; Sheu, I.; Hu, X.; Lin, M.; Zhang, Y.; Chen, Y.-P. Functional Redundancy between HumanSHOXand MouseShox2Genes in the Regulation of Sinoatrial Node Formation and Pacemaking Function. J. Biol. Chem. 2011, 286, 17029–17038.
- Blaschke, R.J.; Hahurij, N.D.; Kuijper, M.S.; Just, S.; Wisse, L.J.; Deissler, K.; Maxelon, T.; Anastassiadis, K.; Spitzer, J.; Hardt, S.E.; et al. Targeted Mutation Reveals Essential Functions of the Homeodomain Transcription Factor Shox2 in Sinoatrial and Pacemaking Development. Circulation 2007, 115, 1830–1838.
- Wang, Q.; Lan, Y.; Cho, E.-S.; Maltby, K.M.; Jiang, R. Odd-skipped related 1 (Odd1) is an essential regulator of heart and urogenital development. Dev. Biol. 2005, 288, 582–594.
- Chiplunkar, A.R.; Lung, T.K.; Alhashem, Y.; Koppenhaver, B.A.; Salloum, F.N.; Kukreja, R.C.; Haar, J.L.; Lloyd, J.A. Krüppel-Like Factor 2 Is Required for Normal Mouse Cardiac Development. PLoS ONE 2013, 8, e54891.
- Gawdzik, J.C.; Yue, M.S.; Martin, N.R.; Elemans, L.M.; Lanham, K.A.; Heideman, W.; Rezendes, R.; Baker, T.R.; Taylor, M.R.; Plavicki, J.S. sox9b is required in cardiomyocytes for cardiac morphogenesis and function. Sci. Rep. 2018, 8, 13906.
- Moskowitz, I.P.; Wang, J.; Peterson, M.A.; Pu, W.T.; MacKinnon, A.C.; Oxburgh, L.; Chu, G.C.; Sarkar, M.; Berul, C.; Smoot, L.; et al. Transcription factor genes Smad4 and Gata4 cooperatively regulate cardiac valve development. Proc. Natl. Acad. Sci. USA 2011, 108, 4006–4011.
- Lindsay, E.A.; Vitelli, F.; Su, H.; Morishima, M.; Huynh, T.; Pramparo, T.; Jurecic, V.; Ogunrinu, G.; Sutherland, H.F.; Scambler, P.J.; et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 2001, 410, 97–101.
- Kume, T.; Jiang, H.; Topczewska, J.M.; Hogan, B.L. The murine winged helix transcription factors, Foxc1 and Foxc2, are both required for cardiovascular development and somitogenesis. Genes Dev. 2001, 15, 2470–2482.
- Bergwerff, M.; Groot, A.G.-D.; Wisse, L.J.; DeRuiter, M.C.; Wessels, A.; Martin, J.F.; Olson, E.N.; Kern, M.J. Loss of function of the Prx1 and Prx2 homeobox genes alters architecture of the great elastic arteries and ductus arteriosus. Virchows Arch. 2000, 436, 12–19.
- Li, R.-G.; Li, L.; Qiu, X.; Yuan, F.; Xu, L.; Li, X.; Xu, Y.; Jiang, W.-F.; Jiang, J.-Q.; Liu, X.; et al. GATA4 loss-of-function mutation underlies familial dilated cardiomyopathy. Biochem. Biophys. Res. Commun. 2013, 439, 591–596.
- Li, J.; Liu, W.-D.; Yang, Z.-L.; Yuan, F.; Xu, L.; Li, R.-G.; Yang, Y.-Q. Prevalence and spectrum of GATA4 mutations associated with sporadic dilated cardiomyopathy. Gene 2014, 548, 174–181.
- Zhao, L.; Xu, J.-H.; Xu, W.-J.; Yu, H.; Wang, Q.; Zheng, H.-Z.; Jiang, W.-F.; Jiang, J.-F.; Yang, Y.-Q. A novel GATA4 loss-of-function mutation responsible for familial dilated cardiomyopathy. Int. J. Mol. Med. 2013, 33, 654–660.
- Zhang, X.-L.; Qiu, X.-B.; Yuan, F.; Wang, J.; Zhao, C.-M.; Li, R.-G.; Xu, L.; Xu, Y.-J.; Shi, H.; Hou, X.-M.; et al. TBX5 loss-of-function mutation contributes to familial dilated cardiomyopathy. Biochem. Biophys. Res. Commun. 2015, 459, 166–171.
- Zhou, W.; Zhao, L.; Jiang, J.-Q.; Jiang, W.-F.; Yang, Y.-Q.; Qiu, X.-B. A novel TBX5 loss-of-function mutation associated with sporadic dilated cardiomyopathy. Int. J. Mol. Med. 2015, 36, 282–288.
- Xu, J.-H.; Gu, J.-Y.; Guo, Y.-H.; Zhang, H.; Qiu, X.-B.; Li, R.-G.; Shi, H.-Y.; Liu, H.; Yang, X.-X.; Xu, Y.-J.; et al. Prevalence and Spectrum of NKX2-5 Mutations Associated With Sporadic Adult-Onset Dilated Cardiomyopathy. Int. Heart J. 2017, 58, 521–529.
- Yuan, F.; Qiu, Z.-H.; Wang, X.-H.; Sun, Y.-M.; Wang, J.; Li, R.-G.; Liu, H.; Zhang, M.; Shi, H.-Y.; Zhao, L.; et al. MEF2C loss-of-function mutation associated with familial dilated cardiomyopathy. Clin. Chem. Lab. Med. 2017, 56, 502–511.
- Li, R.-G.; Xu, Y.; Wang, J.; Liu, X.; Yuan, F.; Huang, R.-T.; Xue, S.; Li, L.; Liu, H.; Li, Y.-J.; et al. GATA4 Loss-of-Function Mutation and the Congenitally Bicuspid Aortic Valve. Am. J. Cardiol. 2018, 121, 469–474.
- Qu, X.-K.; Qiu, X.-B.; Yuan, F.; Wang, J.; Zhao, C.-M.; Liu, X.-Y.; Zhang, X.-L.; Li, R.-G.; Xu, Y.-J.; Hou, X.-M.; et al. A Novel NKX2.5 Loss-of-Function Mutation Associated With Congenital Bicuspid Aortic Valve. Am. J. Cardiol. 2014, 114, 1891–1895.
- Posch, M.G.; Boldt, L.-H.; Polotzki, M.; Richter, S.; Rolf, S.; Perrot, A.; Dietz, R.; Özcelik, C.; Haverkamp, W. Mutations in the cardiac transcription factor GATA4 in patients with lone atrial fibrillation. Eur. J. Med. Genet. 2010, 53, 201–203.
- Yuan, F.; Qiu, X.-B.; Li, R.-G.; Qu, X.-K.; Wang, J.; Xu, Y.-J.; Liu, X.; Fang, W.-Y.; Yang, Y.-Q.; Liao, D.-N. A novel NKX2-5 loss-of-function mutation predisposes to familial dilated cardiomyopathy and arrhythmias. Int. J. Mol. Med. 2014, 35, 478–486.
- Ma, J.-F.; Yang, F.; Mahida, S.N.; Zhao, L.; Chen, X.; Zhang, M.L.; Sun, Z.; Yao, Y.; Zhang, Y.-X.; Zheng, G.-Y.; et al. TBX5 mutations contribute to early-onset atrial fibrillation in Chinese and Caucasians. Cardiovasc. Res. 2016, 109, 442–450.
- Yang, F.; Zhou, L.; Wang, Q.; You, X.; Li, Y.; Zhao, Y.; Han, X.; Chang, Z.; He, X.; Cheng, C.; et al. NEXN inhibits GATA4 and leads to atrial septal defects in mice and humans. Cardiovasc. Res. 2014, 103, 228–237.
- Klattenhoff, C.A.; Scheuermann, J.C.; Surface, L.E.; Bradley, R.K.; Fields, P.A.; Steinhauser, M.L.; Ding, H.; Butty, V.L.; Torrey, L.; Haas, S.; et al. Braveheart, a Long Noncoding RNA Required for Cardiovascular Lineage Commitment. Cell 2013, 152, 570–583.
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Währisch, S.; Beisaw, A.; Macura, K.; Bläss, G.; Kellis, M.; Werber, M.; et al. The Tissue-Specific lncRNA Fendrr Is an Essential Regulator of Heart and Body Wall Development in the Mouse. Dev. Cell 2013, 24, 206–214.
- Ritter, N.; Ali, T.; Kopitchinski, N.; Schuster, P.; Beisaw, A.; Hendrix, D.A.; Schulz, M.H.; Müller-McNicoll, M.; Dimmeler, S.; Grote, P. The lncRNA Locus Handsdown Regulates Cardiac Gene Programs and Is Essential for Early Mouse Development. Dev. Cell 2019, 50, 644–657.e8.




