Cardiac development is a complex developmental process that is initiated soon after gastrulation, as two sets of precardiac mesodermal precursors are symmetrically located and subsequently fused at the embryonic midline forming the cardiac straight tube. Thereafter, the cardiac straight tube invariably bends to the right, configuring the first sign of morphological left–right asymmetry and soon thereafter the atrial and ventricular chambers are formed, expanded and progressively septated. As a consequence of all these morphogenetic processes, the fetal heart acquired a four-chambered structure having distinct inlet and outlet connections and a specialized conduction system capable of directing the electrical impulse within the fully formed heart.
- cardiac development
- congenital heart diseases
1. Introduction
Over the last decades, our understanding of the cellular and molecular mechanisms driving cardiac development has greatly increased. Such discoveries have provided clues to dissect the genetic and molecular bases of congenital heart diseases, as well as provided tools to configure novel cellular and molecular approaches to heal the damaged heart. In this review, we provide a comprehensive summary of the cellular and molecular pathways involved in heart formation, and translational contribution of such findings.
2. From Gastrulation to the Early Cardiac Linear Tube
Cardiac development is complex developmental process that is initiated soon after gastrulation. Soon thereafter the epiblast starts delaminating and migrating towards the future mesodermal layer, and cardiac precursors can be traced in the primitive streak (Figure 1A). At this stage, mesoderm precursors are characterized by the expression of Brachyury [1], Mesp1 and Mesp2 [2][3][4][2,3,4], having all of them an important contribution to early cardiogenic development [5][6][7][8][9] [5,6,7,8,9]. Following the first configuration of the mesodermal layer, cardiac precursors migrate anteriorly and they start expressing early cardiogenic transcription factors. At this stage, several members of the Forkhead, Nkx, Gata, and Mef2 families, respectively, are expressed in the precardiac mesoderm [10][11][12][13][10,11,12,13] in a wide range of different species such as Xenopus [14], zebrafish [15][16][17][15,16,17], chicken [18][19][20][21][22][18,19,20,21,22], and mice [23][24][25][26][27][28][29][30][23,24,25,26,27,28,29,30], being particularly important Nkx2.5 [31][32][33][34][31,32,33,34], Gata4 [35][36][37][38][35,36,37,38], and Mef2c [39][40][41][42][43][39,40,41,42,43] for early cardiogenesis in different experimental models. Expression of these cardiogenic markers is regulated by signaling factors from the adjacent tissues such as Bmp2 [44][45][46][47][48][49][44,45,46,47,48,49], Fgf8 [50][51][52][53][50,51,52,53] and Wnt signaling [54,55,56,57,58,59,60][54][55][56][57][58][59][60]. Importantly, Bmp2 and Fgf8 signaling is controlled by an intricated molecular cascade in which non-coding RNAs, such as miR-130 and miR-133, are also involved [61]. As development proceed, the initial cardiogenic precursors become configured into a horseshoe shape (Figure 1B) and subsequently the bilateral cardiogenic precursors are fused in the embryonic midline generating a cardiac straight tube [62]. At this stage, the embryonic heart is configured by two distinct epithelial layers, an externally located myocardial layer and an internally located endocardial layer. The two distinct cardiogenic subpopulations can be already recognized at this stage [63][64][63,64], the first heart field (FHF) that contributes to the early cardiac straight tube, and the second heart field (SHF) that is medially located and will subsequently contribute to both the arterial and venous poles of the heart [65][66][67][65,66,67] (Figure 1C). FHF specification is dependent of cardiogenic factors such as Nkx2.5 [68], while SHF specification is mostly determined by islet-1 [69] while Bmp signaling contributes to SHF proliferation [70]. On the other hand, Mef2c is required for both FHF and SHF development [71].
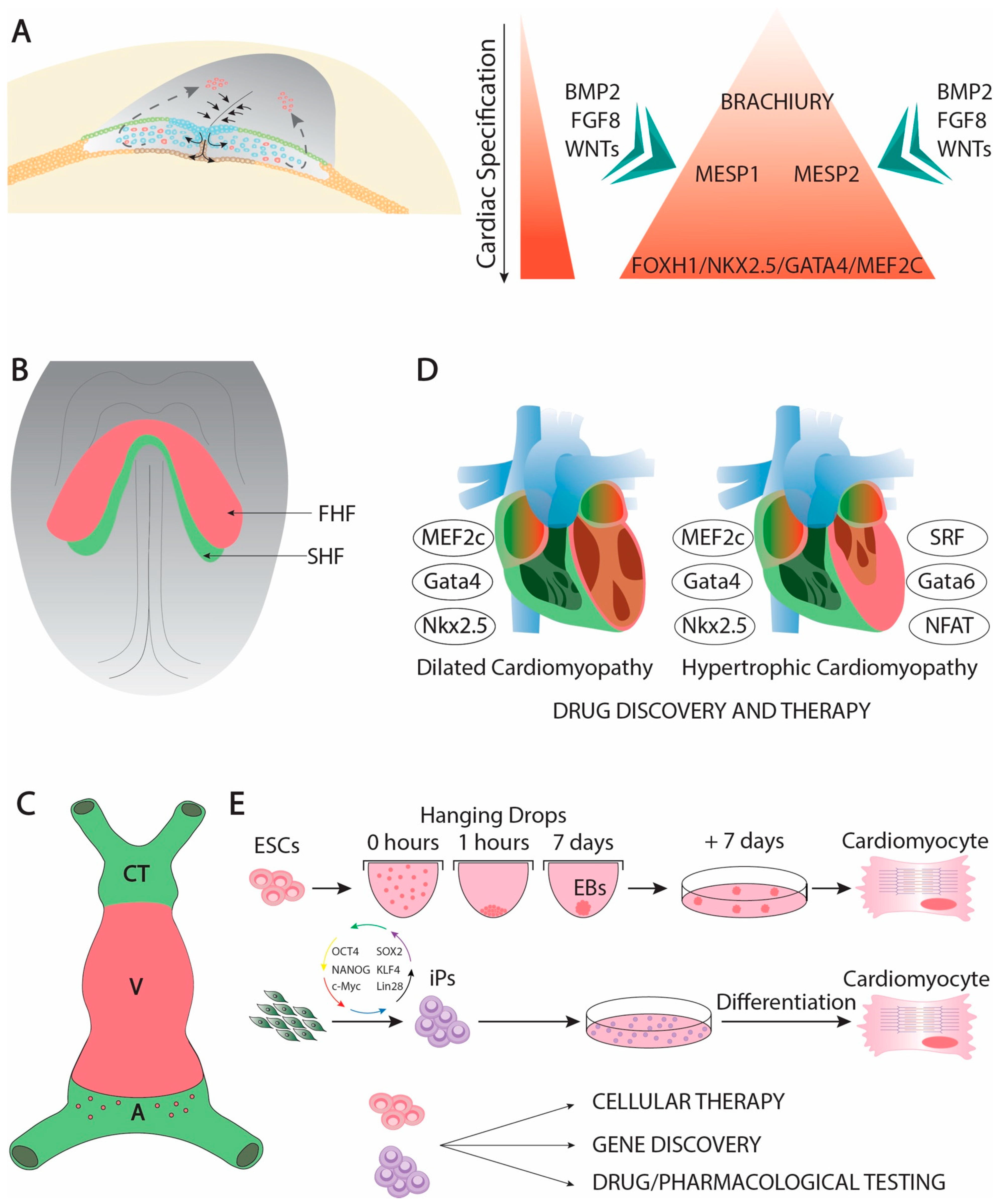
Genetically modified mice have uncovered key functional roles of distinct transcription factors during early cardiac developmental stages. In this context, Gata4 systemic mutants leads to absence of cardiac tube formation [94][95][94,95]. Mef2c and Foxh1 systemic mutants are arrested at the cardiac linear heart tube stage [28][35][28,35], while Nkx2.5 mutants failed to develop right after cardiac looping, providing some cues for ventricular left–right development [96]. Importantly, Mef2c, Gata4, and Nkx2.5 constitute coregulators of early cardiac development [97][98][99][100][97,98,99,100]. Moreover, these core cardiac transcription factors can also be associated with additional cofactors during early cardiogenesis such as Tbx5 [101][102][103][104][101,102,103,104], Gata5 [105], and Gata 6 [106], constituting a complex gene regulatory network [107][108][109][107,108,109] that also implies other transcription factors as reported in different experimental models [110][111][112][113][114][110,111,112,113,114].
These transcriptional networks have been studied in detail over the last decades, uncovering a multiple of downstream genes [115][116][117][118][119][120][121][122][115,116,117,118,119,120,121,122], including non-coding RNAs, such as miR-99/let-7 [123][124][123,124], that are involved in multiple steps of cardiac development as reported in different species. Overall, these data demonstrate that early cardiac development is an intricated morphogenetic mechanisms in which multiple factors are critically involved (Table 1).
Table 1. List of distinct transcription factors involved in cardiogenesis and they main functional contribution to heart development.
| TF | Function | References |
|---|
| Brachyury |
| Mesodermal commitment | [1] | |
| Mesp1 | Cardiogenic mesoderm commitment | |
| Cardiogenic mesoderm commitment | ||
| [ | 2 | ][3][ |
| [ | 2 | |
| 4 | ] | |
| , | 3 | |
| [ | 5 | ] |
| , | 4 | |
| [ | 6 | ] |
| , | 5 | |
| [ | 7 | ][8][9] |
| , | 6,7,8,9] | |
| Mesp2 | Cardiogenic mesoderm commitment | [2][3][4][5][6][7][8][9] |
| Cardiogenic mesoderm commitment | [2,3,4,5,6,7,8,9] | |
| Gata4 | Early cardiac specification, proepicardium development, chamber formation, atrial and atrioventricular septation, cardiomyocyte proliferation, cardiac hypertrophy | [10][11][12][13][14][27][35][36][37][38][94][95][97][98][99][100][125][126][127] |
| Early cardiac specification, proepicardium development, chamber formation, atrial and atrioventricular septation, cardiomyocyte proliferation, cardiac hypertrophy | [10,11,12,13,14,27,35,36,37,38,94,95,97,98,99,100,125,126,127] | |
| Gata5 | Early cardiac specification, cardiomyocyte proliferation | [10][11][12][13][14][16][26][105] |
| Early cardiac specification, cardiomyocyte proliferation | [10,11,12,13,14,16,26,105] | |
| Gata6 | Early cardiac specification, cardiac hypertrophy | [10][11][12][13][14][16][25][41] |
| Early cardiac specification, cardiac hypertrophy | [10,11,12,13,14,16,25,41] | |
| Nkx2.3 | ||
| Early cardiac specification | [15] | |
| Nkx2.5 | Early cardiac specification, FHF development, cardiac looping, chamber formation, cardiac conduction system specification, atrial septation | [15][31][32][33][34][68] |
| [ | 15 | |
| [ | ||
| , | 31 | |
| 96 | ||
| , | 32 | ,33 |
| ] | ||
| , | 34 | |
| [ | 97 | |
| , | 68 | |
| ] | [ | 98 |
| , | 96 | |
| ] | [ | 99 |
| , | 97 | |
| ] | ||
| , | 98 | |
| [ | ||
| , | 99 | |
| 100 | ] | [128][129 |
| , | 100 | |
| ] | [ | 130][131][132][133] |
| , | 128 | |
| [ | ||
| , | 129 | |
| 134 | ||
| , | 130 | |
| ] | [ | 135] |
| Early cardiac specification, FHF development, cardiac looping, chamber formation, cardiac conduction system specification, atrial septation | ,131,132,133,134,135] | |
| Nkx2.6 | Early cardiac specification | [24][29] |
| Early cardiac specification | [24,29] | |
| Nkx2.7 | ||
| Early cardiac specification | [15] | |
| Nkx2.8 | Early cardiac specification | [18][19][20] |
| Early cardiac specification | [18,19,20] | |
| Foxh1 | Anterior heart field development | [28][35] |
| Anterior heart field development | [28,35] | |
| Mef2c | Early cardiac specification, chamber formation | [35][36][37][38][39][40][41][42][43][136] |
| Early cardiac specification, chamber formation | [35,36,37,38,39,40,41,42,43,136] | |
| Islet-1 | ||
| Second heart field specification | [69] | |
| Tbx5 | Early cardiac specification, chamber formation, cardiac conduction system specification, atrial and ventricular septation | [101][102][103][104][ |
| , | 103 | |
| 128 | ||
| , | 104 | |
| ] | ||
| , | 128 | |
| [ | 137 | ][138][139][140] |
| , | 137 | |
| [ | ||
| , | 138 | |
| 141 | ||
| , | 139 | |
| ] | [ | 142] |
| Early cardiac specification, chamber formation, cardiac conduction system specification, atrial and ventricular septation | [101,102,140,141,142] | |
| Pitx2 | left right signalling, heterotaxia, chamber formation, cardiac conduction system development | [143][144][145][146][147][148][149][150][151][152][153][154][155][156][157][158][159][160][161][162][163][164][165][166][167] |
| left right signalling, heterotaxia, chamber formation, cardiac conduction system development | [143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167] | |
| Prrx1 | ||
| cardiac looping | [168] | |
| Wt1 | proepicardium and epicardial development | [169][ |
| proepicardium and epicardial development | ||
| 170 | ] | |
| [ | 169,170] | |
| Tcf21 | proepicardium and epicardial development | [171][172] |
| proepicardium and epicardial development | [171,172] | |
| Tbx18 | ||
| proepicardium and epicardial development | [169] | |
| Tbx2 | primary myocardium and cardiac conduction system development | [173][174][175][176][177][178] |
| primary myocardium and cardiac conduction system development | [173,174,175,176,177,178] | |
| Tbx3 | primary myocardium and cardiac conduction system development | [179][180][181][182] |
| primary myocardium and cardiac conduction system development | [179,180,181,182] | |
| Tbx20 | chamber formation, atrioventricular canal development, cardiomyocyte cell proliferation | [183][184][185][186][187][188][189][190] |
| chamber formation, atrioventricular canal development, cardiomyocyte cell proliferation | [183,184,185,186,187,188,189,190] | |
| Irx3 | ventricular trabecular and cardiac conduction system development | [191][ |
| ventricular trabecular and cardiac conduction system development | ||
| 192 | ] | |
| [ | 191,192] | |
| Irx4 | ventricular chamber development | [191][193] |
| ventricular chamber development | [191,193] | |
| Irx5 | cardiac conduction system development | [191][192] |
| cardiac conduction system development | [191,192] | |
| Hey1 | ventricular chamber development | [194][195][196] |
| ventricular chamber development | [194,195,196] | |
| Hey2 | ventricular chamber development | [194][195][196][197] |
| ventricular chamber development | [194,195,196,197] | |
| Coup-TFII | ||
| atrial development | [198] | |
| eHand | ventricular development | [199][200][201][202] |
| ventricular development | [199,200,201,202] | |
| dHand | ventricular development | [199][202] |
| ventricular development | [199,202] | |
| Foxm1 | ||
| ventricular trabecular and compact layers development | [203] | |
| Hop | ||
| ventricular trabecular and compact layers development | [204] | |
| Klf13 | ||
| ventricular trabecular and compact layers development | [205] | |
| Srf | ventricular trabecular and compact layers development | [206][207] |
| ventricular trabecular and compact layers development | [206,207] | |
| Shox2 | cardiac conduction development | [208][209][210][211][212][213] |
| cardiac conduction development | [208,209,210,211,212,213] | |
| Odd1 | ||
| atrial septation | [214] | |
| Klf2 | ||
| atrioventricular septation | [215] | |
| Sox9 | ||
| atrioventricular septation | [216] | |
| Smad4 | ||
| atrioventricular septation | [217] | |
| Tbx1 | ||
| outflow tract and aortic arch development | [218] | |
| Foxc1 | ||
| aortic arch development | [219] | |
| Foxc2 | ||
| aortic arch development | [219] | |
| Prx1 | ||
| aortic arch development | [220] | |
| Prx2 | ||
| aortic arch development | [220] |
3. Perspectives
Over the last decades, our understanding of the cellular and molecular processes involved in cardiac morphogenesis has greatly increased. A large number of studied in different species have identified several growth factors and transcription factors with pivotal roles in the cardiogenic lineage commitment. Importantly, several of these factors contribute to multiple facets of cardiac development beyond just the early stages, such as for example chamber formation and valvulogenesis as reported for Nxk2.5 and Gata4 ([11][12][13][11,12,13]), respectively. Furthermore, the functional roles of these early cardiogenic lineage transcription factors are also associated to adult cardiac structural pathologies, such as dilated cardiomyopathy [221][222][223][224][225][226][227][420,421,422,423,424,425,426] or bicuspid aortic valves [228][229][427,428] and electrophysiological pathologies such as atrial fibrillation [230][231][232][233][429,430,431,432]. Manipulation of these core transcription factors have provided new tools to convert differentiated cells such as fibroblasts into cardiomyocytes [91], opening new therapeutic opportunities to heal the damaged heart. Importantly, a novel layer of gene regulation is emerging with the identification and functional characterization of non-coding RNAs that are important for cardiomyogenic commitment [234][235][236][462,463,464,465], broadening thus the therapeutic tools to provide novel approaches for cardiac repair.
The embryonic heart is the first organ to display morphological left–right asymmetry. Impairment of sidedness leads to severe body plan abnormalities, including herein cardiac defects. Our knowledge of the molecular and cellular bases of left-right symmetry break has considerably increased over the last decade, providing novel links between early molecular events and impaired sidedness [222]. Such discoveries have served to unravel the genetic bases of heterotaxia and thus as guiding tools for genetic screening and counseling in these human conditions. The discovery of non-coding RNAs involved in early cardiac looping events [252] also exemplify that the final picture of left–right signaling is still incomplete and it would anticipate novel discoveries in the front in the near future.
As cardiac looping takes place, the heart is progressively externally covered by the embryonic epicardium. Importantly, such embryonic epicardium will migrate and deepen into the embryonic myocardium leading to the formation of the cardiac fibroskeleton and part of the coronary vasculature [290,291,292,293]. Failure on the formation of the embryonic epicardium or its progenitor cells in the proepicardium is incompatible with life since the ventricular walls are thinner and the coronary vasculature failed to properly form [299]. Curiously, neither the proepicardium or the epicardium leads to significant contribution to the embryonic myocardium in vivo, but if isolated and cultured in vitro, they can do so [290]. Such properties can be used to unlock the cardiomyogenic potential of the epicardium and thus to sever as a source of myocardial differentiation to heal the heart, as previously reported [300,466]. Unlocking these events with non-coding RNAs have been recently reported [321], opening new strategies to heal the damaged heart.
The formation of the cardiac chambers and the cardiac conduction system is an intricated developmental process that is initiated right after left–right symmetry break and thus cardiac looping. Subsequently, the heart displays a complex septation process that provide morphogenetic cues to form a four-chambered organ with distinct inlet and outlet connections. A multitude of different growth factors such as Bmp, Fgf and Wnt family members, and of transcription factors participate on this cardiac developmental orchestra [44][45][46][47][48][49][50][51][52][53][54][55][56][57][58][59][60][44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60], and similarly as in the earlier events of heart formation, non–coding RNAs, including herein microRNAs and lncRNAs also participate [61][234][235][236][61,315,462,463,464], although our understanding of their functional role is still scarce. Deciphering the molecular cascades involved in these developmental processes have provided candidate genes to test and identify genetic culprits of congenital cardiac anomalies in humans, and thus to design strategies for genetic screening and counseling.
