
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Michail Kotsyfakis | + 972 word(s) | 972 | 2021-01-22 04:58:26 | | | |
| 2 | Lily Guo | Meta information modification | 972 | 2021-02-07 09:54:19 | | | | |
| 3 | Lily Guo | Meta information modification | 972 | 2021-02-07 09:54:45 | | |
Video Upload Options
Long non-coding (lnc)RNAs have emerged as critical regulators of gene expression and are involved in almost every cellular process. They can bind to other molecules including DNA, proteins, or even other RNA types such messenger RNA or small RNAs. LncRNAs are typically expressed at much lower levels than mRNA, and their expression is often restricted to tissue- or time-specific developmental stages. They are also involved in several inter-species interactions, including vector–host–pathogen interactions, where they can be either vector/host-derived or encoded by pathogens. In these interactions, they function via multiple mechanisms including regulating pathogen growth and replication or via cell-autonomous antimicrobial defense mechanisms. Recent advances suggest that characterizing lncRNAs and their targets in different species may hold the key to understanding the role of this class of non-coding RNA in interspecies crosstalk.
1. Introduction
LncRNAs are therefore a diverse group of regulatory ncRNAs with various characteristics, localizations, and modes of action[1]. A comprehensive classification of lncRNAs is quite challenging, as most do not share structural, functional, or mechanistic features. Moreover, only a few lncRNAs have been thoroughly mechanistically characterized to date, with even fewer being functionally verified in vivo[2]. Overall, their functions depend on their subcellular localization, i.e., in the cytoplasm or nucleus. Here we describe the five archetypal molecular functions of lncRNAs: as signals, decoys, guides, scaffolds, and enhancers (Figure 1).
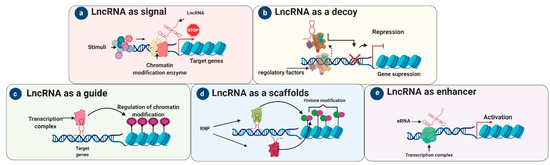
Figure 1. Archetypes and mechanisms of action of lncRNAs. LncRNAs execute five main molecular functions depending on their mechanism of action. (a) Signal lncRNAs: responding to a stimulus, lncRNAs receive the signal to interact with chromatin-modifying enzymes to prevent transcription. (b) Decoy lncRNAs have more affinity for particular regulatory factors; once bound, they lead to transcriptional repression by preventing these regulatory factors from binding to the DNA. (c) Guide lncRNAs arrange transcription factors at specific genomic loci and help to regulate chromatin. (d) Scaffold lncRNAs help to assemble RNA-protein (RNP) complexes; their role is to promote or suppress transcription by activating or repressing target genes. (e) Chromatin interactions are influenced by an enhancer that can be presented by an enhancer RNA (eRNA).
2. LncRNAs as Signals
Signal lncRNAs are expressed at a specific time and in a specific position in the cell in response to stimuli. Some lncRNAs in this archetype are regulatory, while others are merely by-products of transcription, with the act of initiation, elongation, or termination being regulatory. Signal lncRNAs (Figure 1) are known to interact with chromatin-modifying enzymes such as histone methyltransferases to silence their target genes by blocking their transcription or through heterochromatin formation [3].
3. LncRNA as Decoys
“Decoy” lncRNAs can act as molecular sinks by decreasing the accessibility of particular regulatory factors via decoy binding sites[3]. These lncRNAs repress transcription, in an indirect way, by sequestering regulatory factors (Figure 1) including miRNAs, modifying complexes (chromatin subunits), catalytic proteins, and transcription factors, reducing their availability [3][4]. LncRNA decoys negatively modulated transcription by not allowing a particular effector to interact with its intrinsic target [3]. For example, the decoy lncRNA PANDA regulates apoptosis [3]: NF-YA is a transcription factor responsible for the activation of apoptosis-related genes, but when PANDA binds to NF-YA it is sequestered away from its target genes, resulting in the suppression of apoptosis-related genes[5][6]. Other decoy lncRNAs like TUG1 and MEG3 indirectly degrade and alter protein translation by isolating miRNA from mRNA and protein targets[3][7][8]. In addition, many other lncRNAs, such as highly up-regulated in liver cancer (HULC), H19, growth arrest-specific 5 (GAS5), HOTAIR, phosphatase and tensin homolog pseudogene 1 (PTENP1), and MALAT1, act as decoys by preventing transcription factors or miRNA from binding with their target sites, thereby regulating translation and transcription (Figure 1).
4. LncRNA as Guides
To regulate the genome, guide lncRNAs are necessary for the organization/localization of factors at specific genomic loci. As such, lncRNA guides direct chromatin modifiers and protein complexes (such as RNPs) or transcription factors to particular target gene(s) and help them localize appropriately at their transcriptional loci [9]. The exact targets of guide lncRNAs (Figure 1) are stimulated by RNA–RNA, RNA–protein, and RNA–DNA interactions. However, the exact details of this mechanism are uncertain [10]. LncRNAs such as HOTAIR, functional intergenic repeating RNA element (FIRRE), COLDAIR, KCNQ1 overlapping transcript 1 (KCNQ1OT1), taurine-upregulated gene 1 (TUG1), HOTTIP, XIST, MEG3, and ANRIL can act as guides by reprograming chromatin complexes and managing the recruitment of epigenetic modifiers to their definitive loci[11]. For instance, KCNQ1OT1 and HOTAIR bind to PRC2, a chromatin modifier, to regulate target genes in cis or trans and thereby inhibit gene expression [12][13][14].
5. LncRNA as Scaffolds
Scaffold lncRNAs play an important structural role in assembling multi-protein complexes such as short-lived ribonucleoprotein (RNP) complexes[15]. After the complete assembly of RNP complexes, they can suppress or activate transcription depending on the existence and nature of the involved RNAs and proteins[15]. For example, the well-studied lncRNA TERC is an excellent example of a molecular scaffold, in which the telomerase complex combines associated proteins with reverse transcriptase action in a single RNP[16]. TERC is a more stable molecular scaffold lncRNA than all other newly identified lncRNAs (Figure 1). As substitutes, lncRNAs might have low-affinity interactions with protein-like mRNAs as they mature. In addition, lncRNAs like MALAT1, ANRIL, and TUG1 can serve as dynamic scaffolds by associating with PRC1 and PRC2 to stimulate target gene suppression or activation[4][17].
6. LncRNA as Enhancers
In the signal archetype, the lncRNA sends a molecular signal to initiate regulation at the transcriptional level in response to different stimuli [8][18]. For instance, KCNQ1OT1 recruits PRC2 and chromatin-modifying enzymes (histone methyltransferases) by acting as a signal lncRNA [19]. In the enhancer archetype (Figure 1), the “chromatin interaction” of DNA is influenced by the enhancer regions (ERs) as a result of enhancer RNAs (eRNAs). It is thought that these lncRNAs are not released from ERs but tether proteins to ERs [20].
Finally, lncRNAs may have additional regulatory functions such as trafficking and protein signaling, which require further study.
References
- Kung, J.T.Y.; Colognori, D.; Lee, J.T. Long noncoding RNAs: Past, present, and future. Genetics 2013, 193, 651–669.
- Bassett, A.R.; Akhtar, A.; Barlow, D.P.; Bird, A.P.; Brockdorff, N.; Duboule, D.; Ephrussi, A.; Ferguson-Smith, A.C.; Gingeras, T.R.; Haerty, W.; et al. Considerations when investigating lncRNA function in vivo. Elife 2014, 3, 1–14, doi:10.7554/eLife.03058.
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914.
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene 2011, 30, 1956–1962, doi:10.1038/onc.2010.568.
- Hung, T.; Wang, Y.; Lin, M.F.; Koegel, A.K.; Kotake, Y.; Grant, G.D.; Horlings, H.M.; Shah, N.; Umbricht, C.; Wang, P.; et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011, 43, 621–629, doi:10.1038/ng.848.
- Baldassarre, A.; Masotti, A. Long Non-Coding RNAs and p53 Regulation. Int. J. Mol. Sci. 2012, 13, 16708–16717, doi:10.3390/ijms131216708.
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283, doi:10.1038/nrg.2016.20.
- Pandey, R.R.; Mondal, T.; Mohammad, F.; Enroth, S.; Redrup, L.; Komorowski, J.; Nagano, T.; Mancini-Dinardo, D.; Kanduri, C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 2008, 32, 232–246, doi:10.1016/j.molcel.2008.08.022.
- Chen, J.; Wang, H.; Yao, Y. Experimental study of nonlinear ultrasonic behavior of soil materials during the compaction. Ultrasonics 2016, 69, 19–24, doi:10.1016/j.ultras.2016.03.001.
- Vance, K.W.; Ponting, C.P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014, 30, 348–355, doi:10.1016/j.tig.2014.06.001.
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Währisch, S.; Beisaw, A.; Macura, K.; Bläss, G.; Kellis, M.; Werber, M.; et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 2013, 24, 206–214, doi:10.1016/j.devcel.2012.12.012.
- Davidovich, C.; Cech, T.R. The recruitment of chromatin modifiers by long noncoding RNAs: Lessons from PRC2. Rna 2015, 21, 2007–2022, doi:10.1261/rna.053918.115.
- Lee, J.T. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat. Rev. Mol. Cell Biol. 2011, 12, 815–826, doi:10.1038/nrm3231.
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.-C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076, doi:10.1038/nature08975.
- Yang, L.; Froberg, J.E.; Lee, J.T. Long noncoding RNAs: Fresh perspectives into the RNA world. Trends Biochem. Sci. 2014, 39, 35–43, doi:10.1016/j.tibs.2013.10.002.
- Zappulla, D.C.; Cech, T.R. Yeast telomerase RNA: A flexible scaffold for protein subunits. Proc. Natl. Acad. Sci. USA 2004, 101, 10024–10029, doi:10.1073/pnas.0403641101.
- Yap, K.L.; Li, S.; Muñoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.-M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 2010, 38, 662–674, doi:10.1016/j.molcel.2010.03.021.
- Luisa Pedroso Ayub, A.; D’Angelo Papaiz, D.; da Silva Soares, R.; Galvonas Jasiulionis, M. The Function of lncRNAs as Epigenetic Regulators. Non-Coding RNAs 2020, 2020, doi:10.5772/intechopen.88071.
- Kim, D.-H.; Xi, Y.; Sung, S. Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet 2017, 13, e1006939, doi:10.1371/journal.pgen.1006939.
- Hou, Y.; Zhang, R.; Sun, X. Enhancer lncrnas influence chromatin interactions in different ways. Front. Genet. 2019, 10, 936, doi:10.3389/fgene.2019.00936.




