1000/1000
Hot
Most Recent

Glycans are major constituents of extracellular vesicles (EVs). Alterations in the glycosylation pathway are a common feature of cancer cells, which gives rise to de novo or increased synthesis of particular glycans. Therefore, glycans and glycoproteins have been widely used in the clinic as both stratification and prognosis cancer biomarkers. Interestingly, several of the tumor-associated glycans have already been identified in cancer EVs, which constitutes valuable sources of cancer biomarkers. Furthermore, glycans have also shown to play a role in EV protein sorting, uptake and tropism. Altogether, the EV glycan signatures hold tremendous potential to be applied into the clinical setting for both biomarker discovery and as therapeutic delivery systems.
Extracellular vesicles (EVs) are small nano-sized particles, secreted by all cell types and capable of encapsulating and transporting several molecules to a target delivery site [1]. EVs can be found in various biological fluids and can be harvested in relatively non-invasive ways. Therefore, these particles are attractive systems for targeted drug delivery approaches and valuable sources of circulating cancer biomarkers.
Alterations of the glycosylation pathway are a common feature of malignant cell transformation [2][3][4]. These carbohydrates are capable of modulating several processes during cancer progression, including activation of oncogenic signaling pathways, interference with cell–cell and cell–extracellular matrix (ECM) adhesion and mediate cancer cell metastasis [3][4]. In addition, alteration in the glycosylation pattern of a cell has also been associated with content sorting processes [5][6][7], and with the capacity of cells to interact and uptake certain EVs [8][9][10][11]. Interestingly, some of the tumor-associated glycan alterations have already been identified as enriched in cancer EVs (Figure 1), which may constitute important biomarkers with the potential to be used in the clinical setting.
Figure 1. Schematic representation of an extracellular vesicle (EV) and its functional cargo. EVs carry a wide variety of functional molecules, including glycoproteins and glycolipids. The major common classes of glycoconjugates found in human cells are depicted on the left. Aberrant tumor-associated glycosylation already identified in cancer EVs are depicted on the right. The glycostructures were represented at the expected EV localization (intern or at the EV membrane) considering the knowledge from the cell glycans. Nevertheless, the specific localization of some of these structures is not yet fully elucidated.
Despite technological advances, the structural characterization of glycans remains quite challenging. The diversity and complexity of these carbohydrates, together with methodological limitations, makes it challenging to deeply analyze the EV glycome [12][13]. The presence of specific glycosylation profiles in tumor EVs highlights its potential to be used not only to develop novel cancer EV detection and isolation methods but also as a source of novel circulating biomarkers. In this review, after a brief description of the main types of changes in glycosylation found in cancer and their impact on different pathological processes, we pointed out the challenges faced by the currently available methods used in the analysis of the EVs glycome. In addition, we summarized the glycosylation patterns already identified in tumor-EVs and discussed their known function in cancer and how they have been used to develop additional EV detection and isolation technologies.
Glycans are carbohydrate structures that modify both proteins and lipids through a biosynthetic pathway finely regulated by glycosyltransferases and sugar transporters. Glycans can be mainly found at the surface of the cellular membrane forming the commonly known glycocalyx, and they are essential mediators of cell–cell communication and cell–matrix interaction [3][14][15]. The major types of glycosylation that can be affected in cancer include (I) N-glycans, characterized by an N-linkage to an Asn residue in an Asn-X-Ser/Thr sequon, where X can be any amino acid except proline. N-glycans have a defined core structure, and can be classified depending on their structures and branches in high-mannose, complex or hybrid [16]; (II) O-GalNAc glycans, also known as mucin-type O-glycans, are carbohydrate chains initiated by a GalNAc sugar covalently linked by an oxygen atom to a Ser/Thr residue. They often appear as long ramified structures, and have multiple core structures, the major includes core-1 to core-4 [17]; and (III) glycosaminoglycans (GAGs), long and non-ramified carbohydrate chains consisting of repeating disaccharide units [18]. The glycosylation pathway is highly regulated by numerous players, including the expression, localization and activity of both glycosyltransferases and glycosidases, and the availability of nucleotide sugar donors [19]. Therefore, variations in the expression levels of specific glycosyltransferases [20] or its mislocalization in the endoplasmic reticulum (ER) and Golgi apparatus (GA) [21], dysregulation of chaperone activity [22] or alterations in nucleotide sugar transporter availability and cofactors [19] will result in the synthesis of aberrant glycosylation in cancer.
These macromolecules play pivotal roles in several physio- and pathological processes either by functioning as structural scaffolds, recognition cues or modulators of other important biomolecules [14]. Genetic and epigenetic alterations that disturb the glycosylation machinery often arise during malignant transformation, which results in loss or increased expression of certain glycans and the appearance of novel glycans [2][3]. The presence of aberrant glycosylation in cancer cells can impact several biological processes including tumor cell proliferation [23], angiogenesis [24], invasion [25] and metastasis [26]. Alterations in the glycosylation pathway provide multiple adaptive advantages, including receptor tyrosine kinase (RTK) activation [25][27][28][29], regulation of adhesion-related proteins [30][31][32] and immune response modulation [15][33], which significantly contribute to cancer progression. All these aspects will be briefly discussed, as they have already been extensively reviewed [3][4][34].
EVs are small nano-sized particles that are released into the extracellular space by all types of cells. These vesicles exert a broad array of biological functions, being important mediators of intercellular communication [1]. EV diameter typically ranges from 35–5000 nm, and therefore are quite smaller than cells, but much larger than proteins [35][36]. The general term EVs comprises three main types of vesicles which are classified, according to their size and biogenesis mechanism, into exosomes, microvesicles (ectosomes or microparticles) and apoptotic bodies. Exosomes have an endocytic origin and are produced by the inward budding of the plasma membrane of the cell. This invagination of the cell membrane leads to the formation of multivesicular bodies (MVBs) that can later either fuse with lysosomes for content degradation or fuse with the cellular membrane to be secreted in the form of exosomes. The formation and release of exosomes can be regulated by the endosomal sorting complex required for transport (ESCRT) [37][38]. In this type of vesicles, it is expected to find ESCRT proteins and accessory proteins for this complex, such as the ALG-2-interacting protein X (Alix), the tumor susceptibility gene 101 (TSG101) and the chaperones HSP70, Hsc70 and HSP90β, independently of the type of cell origin [36][39][40]. Another mechanism, independent of the ESCRT complex, can be used for exosome release [41]. In the absence of this complex, the endosome pathway may be regulated by the type II neutral sphingomyelase and the tetraspanin family proteins [42]. Therefore, exosomes will contain high levels of tetraspanins such as CD9, CD63 and CD81 [43]. On the other hand, microvesicles are released into the extracellular space by direct shedding of the plasma membrane [37][38]. Therefore, microvesicles can carry both cytosolic and plasma membrane proteins, including the same tetraspanins found on exosomes [44]. Microvesicles can also contain cytoskeletal and heat shock proteins and integrins [45][46]. Finally, apoptotic bodies are formed during the cellular apoptotic process. The process of cell apoptosis is characterized by the condensation of chromatin followed by the degradation of the internal structure of a cell [37][38]. The disintegrated cellular content will be part of the apoptotic bodies’ cargo. Therefore, this type of vesicles can contain proteins associated with several organelles such as histones (nucleus), the heat shock protein HSP60 (mitochondria) and the chaperone GRP78 (endoplasmic reticulum) [47][48][49]. EVs are composed of a phospholipid bilayer that provides protection to their cargo against degradation by the proteases and nucleases present in the external environment [50][51]. EVs encapsulate several molecules, including cytosolic and cytoskeletal proteins as well as enzymes and nucleic acids (mRNA, miRNA, tRNA, rRNA, DNA) [52][53][54]. The surface of the EVs is composed of lipids (ceramide, cholesterol, phosphatidylserine and sphingomyelin) and proteins (transmembrane proteins, antigen presenters and adhesion molecules) [1]. In addition, glycans are also relevant constituents of the EV composition surface [6][55][35][56].Cancer EVs are able to mediate communication between cells locally and at a distance, and their cargo can influence the behavior of the recipient cell [57]. Importantly, tumor microenvironment stressors, such as hypoxia [58][59], acidosis [60], starvation [61][62], oxidative stress [63][64], radiation [59] and anti-cancer therapies [65], are important regulators of not only EV secretion and trafficking, but also of its molecular composition (as reviewed in [66]). EV cargo is mainly similar to the composition of the parental cell [1]. Although, they still have unique molecular profiles resultant from specific sorting mechanisms during the EV biogenesis process. Particularly, specific patterns of glycans were found enriched in EVs [5][6][67]. In cancer, important modifications occurring in surface glycans, both at cellular and EV level, may constitute important markers for EV detection, isolation and, importantly, for tumoral and non-tumoral EV distinction.
EVs are capable of carrying several bioactive molecules and, depending on their surface composition, hold the potential to be used as natural vehicles for localized drug delivery [68][69]. In addition, as cancer EVs can be found in several biofluids [70] and their cargo partially reflects the content of parental cells [1], these vesicles are also considered promising sources of circulating cancer biomarkers.
EVs can be re-engineered to carry specific molecules, either by manipulating their parental cells or by direct functionalization of the EVs [71][72]. Several studies have shown the possibility of using glycosylphosphatidylinositol (GPI) as an anchor to attach specific antigens to the membrane of EVs for target therapy purposes. For example, the HER-2 remained stable after its fusion with GPI and incorporation into murine breast cancer EVs, which induced strong HER-2-specific antibody responses when injected into mice [73]. In addition, fusing anti-EGFR nanobodies to the GPI anchor of neuroblastoma EVs resulted in a significantly increased capacity of these EVs to bind tumor cells that overexpress the EGFR [74]. Furthermore, the incorporation of the GPI-anchored immune-stimulatory molecule interleukin 12 (IL-12) in EVs isolated from different tumor cell lines resulted in increased in vitro T cell proliferation [75]. These studies highlight the potential of modifying the EV surface to successfully transport antigens to their destination site (Figure 2). To explore the real potential of EVs for cancer drug delivery purposes, a reliable system capable of tracking both in vitro and in vivo interactions of these natural nanoparticles is required. Interestingly, a new method of natural particle labeling based on glycan trafficking was recently reported. In this study, azido-sugars were metabolically incorporated into the cellular glycans and further packaged into the EVs, which allowed these EVs to be traceable in vivo [76].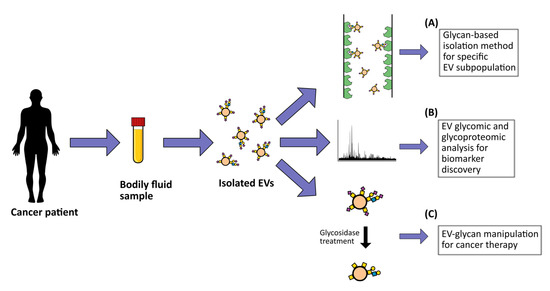
Figure 2. Potential clinical applications of extracellular vesicle (EV) glycosylation. These include (A) development of glycan-based EV detection and capture methodologies; (B) EV biomarker discovery for cancer diagnostic, prognostic and/or patient stratification based on EV glycomic and glycoproteomic profiling; (C) development of potential novel cancer therapy strategies through the manipulation of EV glycosylation surface.After reaching their target destination, EVs can be incorporated by recipient cells and release their content.
As discussed before, the presence of HSPGs on recipient cells has proved to act as receptors for cancer-derived EVs [9]. The structure and function of these HSPGs can be regulated by heparanase [77]. Interestingly, the use of heparanase inhibitors showed to block tumor progression by reducing exosome uptake by receptor cells [11][78][79]. Currently, there are some heparanase inhibitors, such as modified heparins or HS mimetics, whose potential use in the clinic is being tested [80][81][82][83][84]. Examples include chemically modified N-desulfated, N-acetylated and glycol-split heparin derivatives [85] and a heparanase inhibitor [86], that alone or together with lapatinib, resulted in inhibition of the tumor growth in patients with myeloma [82] or in brain metastatic breast cancer, respectively [87]. Interestingly, it was demonstrated that a specific EV glycosylation coating per se can induce a host immunogenic response. In melanoma, the modification of apoptotic EVs surface towards overexpression of high mannose type glycans, a natural ligand of DC-SIGN, increased the uptake of these EVs by monocyte-derived dendritic cells, leading to an increase in CD8+ T cell response [88]. In addition, the enzymatic removal of sialic acids and insertion of palmitoyl-LeY in glioblastoma EVs led to an enhanced EV uptake by dendritic cells in a DC-SIGN dependent manner, a receptor involved in the activation of CD8+ and CD4+ T cell responses [89]. Therefore, these studies showed that modifications of the EV glycan surface hold potential as a vaccination strategy to potentiate an anti-tumor immune response. The capacity of certain EV glycans to naturally stimulate the immune system should be further explored for the development of novel potential immune-related therapies.A different strategy with the potential to be applied for cancer therapy involves the hemofiltration of the patient circulating exosomes using the Aethlon ADAPT™ system (adaptive dialysis-like affinity platform technology) [90]. This system aims to capture tumoral EVs through interaction with their surface proteins or glycans. The efficacy of the ADAPT™ system was evaluated in patients with end-stage renal disease. It was possible to reduce the circulating hepatitis C virus by targeting the high mannose glycans present on the viral particles [91]. Although there is no concrete data on its usage in removing EVs from the circulation of cancer patients, the ADAPT™ system is a promising strategy to capture tumor EVs based on their glycosylation profile.The role of EVs in modulating therapeutic resistance has already been reported (as reviewed in [92]). EVs can be used by tumor cells as resistance mechanisms through the packaging and release of drugs by these vesicles [93][94][95]. Furthermore, the transfer of proteins, such as the multidrug resistance P-glycoprotein [96][97][98][99], or specific microRNAs [100][101][102] from drug-resistant cells to drug-sensitive cells can lead to the modulation of gene expression and the acquisition of resistance in recipient cells. Since glycans present on the EV surface are important mediators of the interaction and uptake of these vesicles by the recipient cells [8][9][10][11], changes in the EV glycosylation will alter the intercellular communication between resistant and sensitive cells. EVs are also capable of modulating the immune response. Through the delivery of specific cargo, such as immune-stimulatory or immune-suppressive molecules, EVs can regulate the activity of immune cells (as reviewed in [103][104]). As previously addressed in this review, glycosylation is an important modulator of the immune response, and cancer cells use specific glycan profiles to escape immunosurveillance. Thus, it is possible that tumor-derived EVs carrying these glycan signatures will also suppress the immune response.
Besides their potential application for cancer therapy, EVs also represent a valuable source of circulating biomarkers (Table 1). High EV concentrations have been found in several body fluids, including blood, urine, saliva, cerebrospinal fluid, lymph, pleural effusions, semen, bronchoalveolar lavage, bile, synovial fluid, nasal secretions, breast milk, ocular effluent and ascites (as reviewed in [105]). As alterations in cell glycosylation are a common feature of cancer progression, and glycans are highly present in cancer EVs [106][107], the disclosure of cancer EV glycosylation holds a tremendous potential to identify novel reliable cancer EV biomarkers. In fact, most of the currently available cancer biomarkers are based on the detection of glycans, including the sialyl Lewis A antigen (CA19-9) and the STn (CA72-4), or glycoproteins, such as the alpha-fetoprotein (AFP), the prostate-specific antigen (PSA), mucin 16 (CA125), mucin 1 (CA15-3) and the carcinoembryonic antigen (CEA), which are used to follow both patient treatment response and tumor recurrence in several types of cancers (as reviewed in [3][4][108]).Interestingly, the carbohydrate antigen CA125 was identified in serum-derived exosomes from patients with ovarian cancer and the detected levels were significantly higher in the exosomes when compared to the levels detected directly in the serum of these patients [109]. Yokose and his collaborators also studied the glycan profiles of serum EVs and revealed a significant increase in O-glycosylated EVs in pancreatic cancer patients in the early stages of the disease, even when the patient samples were negative for the CA19-9 antigen [110]. Interestingly, elevated levels of CA19-9 were detected in exosomes from pancreatic cancer patients when compared to healthy samples. The analysis of CA19-9 in exosomes proved to be more sensitive than its direct measurement from the serum, which allowed to identify CA19-9 positive exosomes in patients thought to be negative for the presence of this antigen [111]. In addition, a highly glycosylated form of the CD133 glycoprotein carrying increased levels of sialic acids was found in exosomes from pancreatic cancer patient’s ascites and was also associated with patient survival. Although further studies are needed, these results demonstrate the prognosis potential of CD133-specific glycosylation in pancreatic cancer [112]. In addition to glycoproteins, the glycosphingolipids abundantly present on the surface of prostate cancer EVs have also been reported as promising biomarkers for this type of cancer [113]. The overexpression or de novo synthesis of particular glycans or glycoconjugates during cancer progression holds the potential to differentiate tumor EVs from benign EVs. Indeed, the proteoglycan glypican-1 (GPC1) and the tumor antigen chondroitin sulfate proteoglycan 4 (CSPG4) were detected in tumor exosomes from heterogeneous samples of pancreatic cancer [114] or melanoma [115], respectively. In both cases, these glycoproteins were able to differentiate tumor-derived EVs from non-malignant particles. In a recent study, the proteoglycan versican (VCAN) and the glycoprotein tenascin C (TNC) also proved to be able to distinguish tumor from non-tumor tissues with high sensitivity and specificity, pointing to their use as cancer EVs markers [116]. In the same study, the galactoside-binding soluble 3 binding protein (LGALS3BP) was identified in most of the EV samples [116], which is in line with the previous reports of the presence of LGALS3BP in uveal melanoma [117] and ovarian cancer EVs [56][118]. Interestingly, the LGALS3BP protein was also found to be strongly enriched in the recently discovered cancer exomere particles [35]. Moreover, the glycoprotein basigin (CD147 or EMMPRIN) and the proteoglycan biglycan (BGN) were found enriched in pancreatic tumor EVs when compared to EVs secreted by non-tumor adjacent tissues [116]. In accordance, highly glycosylated variants of EMMPRIN were predominantly detected on cancer patient-derived microvesicles and were positively correlated with poor survival in several types of cancer [119]. In addition, the O-GlcNAc glycosylation has also been found elevated in breast [120] and colorectal cancer EVs [121], when compared to normal conditions. In particular, the O-GlcNAc modification of the transitional endoplasmic reticulum ATPase (TER ATPase) and RuVB-like1 proteins was identified in colorectal metastatic EVs [121]. Elevated levels of O-GlcNAc were also detected in TER ATPase as well as in 70 kDa heat-shock protein (HSP70) proteins present in breast cancer EVs, which may act to protect cytosolic and nuclear proteins against degradation [120]. The elevated presence of this type of glycosylation modification conjugated with specific proteins identified in tumoral EVs when compared to normal conditions suggests its potential use as a biomarker for both breast cancer and metastatic colorectal cancer.Despite the high potential of EV glycosylation in the discovery of novel cancer biomarkers, studies addressing the glycan profile of blood circulating EVs are still quite scarce. Nevertheless, the N-glycome of exosomes from hepatocellular carcinoma patient samples was characterized using a reverse capture strategy, and the majority of the N-glycans found in EVs from patients with HCC were modified with sialic acids or fucoses, in contrast to the N-glycans identified in EV from healthy samples [122]. Walker et al. also reported significant differences between the glycan profiles identified directly in the plasma or the plasma-derived EVs from the same individuals [123]. Interestingly, a new integrated analytical platform, termed the integrated magnetic analysis of glycans in extracellular vesicles (iMAGE), was developed to directly analyze the EV glycosylation profile in biological samples. This platform aims to facilitate the EV glycome analysis taking advantage of the magnetic nanotechnologies [124]. The effectiveness of this strategy was evaluated by spiking kidney and brain cancer-EVs into urine and serum EV-depleted samples, respectively, and analyzing the glycan signatures. This strategy proved to be efficient in detecting EVs. Subsequently, when analyzing the glycan profile of ascites samples from patients with gastric and colorectal cancer, it was possible to distinguish patients based on their prognosis only by the glycans present at the EV surface. The distinction of these patients was possible by the increased signal of different lectins associated with a poor prognosis, including the Jacalin, ConA, RCA120, PHA-E, STA, LEL, WGA, DSL and LCA lectins. Although further studies are needed to prove the iMAGE platform’s robustness, this new method of profiling glycans may prove to be very useful in the search for novel biomarkers in cancer research [124]. Although several techniques can be used to analyze glycans, their study faces several technical challenges. The most commonly used methods only provide relative and not absolute quantification of the glycans present in a sample and are often based on a targeted search for specific patterns of cancer-associated glycans [125]. In addition, the variability of the results obtained when the same samples are analyzed in different laboratories, in which different methods were used, demonstrates the difficulty in choosing the best methodology, and the need for reference standards that support that choice [126]. Nevertheless, the presence of different glycosylation profiles under normal and cancer conditions highlights the potential of studying EV-specific glycosylation for the identification of novel cancer circulating biomarkers. Indeed, the studies addressing EV glycosylation denote a high potential of EV glycans to distinguish from normal vs. tumoral EVs. Therefore, we believe further in-depth studies of EV glycosylation will bring several benefits for the future of cancer patients.