| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jean Pierre Etchegaray | + 2520 word(s) | 2520 | 2020-12-08 08:59:53 | | | |
| 2 | Rita Xu | -893 word(s) | 1627 | 2020-12-15 06:56:58 | | | | |
| 3 | Rita Xu | Meta information modification | 1627 | 2020-12-16 02:33:23 | | |
Video Upload Options
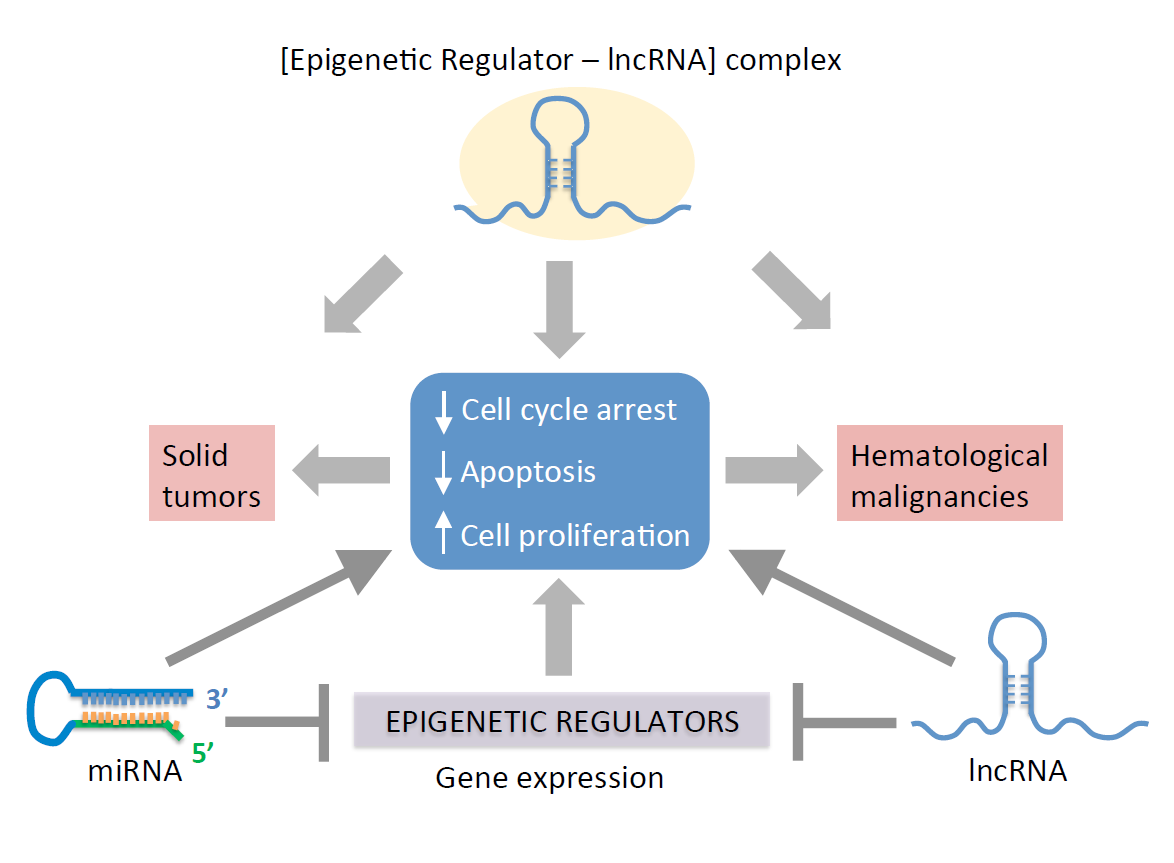
This entry discuss the role of ncRNAs as mediators of epigenetic pathways associated with carcinogenesis, and provides an overview on how deregulated microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) alter the expression of cancer-promoting genes. Overall, miss regulated expression of ncRNAs alter cell cycle progression and apoptosis to increase proliferation, thereby facilitating cellular transformation into malignancies. The potential for using ncRNAs as targets for early prognosis and for developing cancer therapies is discussed.
1. Introduction
The pathophysiology of cancer is associated with multiple molecular and cellular dysfunctions, including genetic and/or epigenetic alterations [1]. Epigenetic changes are catalyzed by specific enzymes capable of modifying the chromatin structure, e.g., acetylation, methylation, phosphorylation, ubiquitylation and glycosylation [2][3]. These modifications can be influenced by environmental changes [4][5]. The cellular epigenetic landscape can be regulated by non-coding RNAs (ncRNAs) to modify gene expressions [6][7][8]. ncRNAs are untranslated RNA molecules capable of regulating gene expression through multiple pathways. For instance, ncRNAs can target the chromatin to induce gene silencing through direct interaction with epigenetic factors. ncRNAs can also interact with transcription factors to either prevent or promote the expression of target genes. Additionally, ncRNAs can silence gene expressions by directly binding to mRNA targets, a process known as RNA-induced silencing. This review article is focused on the epigenetic function of small and long ncRNAs: microRNAs (miRNAs), short interfering RNAs (siRNAs) and long non-coding RNAs (lncRNAs) in the context of cancer biology.
In general, miRNAs typically repress specific gene targets by RNA-induced silencing at the post-transcriptional level [9]. In contrast, lncRNAs can use multiple methods to regulate gene expressions. These include the remodeling of chromatin to activate or repress transcription, modulating pre-mRNA splicing and inhibiting mRNA translation [10]. In addition to miRNAs and lncRNAs, other ncRNAs have specialized functions. These include small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), ribosomal RNAs (rRNAs), transfer RNAs (tRNAs) and piwi-interacting RNAs (piRNAs). snRNAs are approximately 150 nucleotide bases and are primarily located within the splicing regions of nuclei. Due to their locations, snRNAs have been reported to be associated with pre-mRNA splicing, particularly in the formation of the spliceosome [11]. The spliceosome is a large, dynamic complex that is composed of the five snRNAs: U1, U2, U4, U5 and U6, along with other protein components [11][12][13]. The combination of snRNAs and other splicing proteins, a small nuclear ribonucleoprotein or snRNP complex, includes a singular spliceosomal snRNA within a complex with various splicing proteins [11]. Similar to snRNAs are snoRNAs; there is documentation of a group of ncRNAs primarily located in the nucleoli of eukaryotic cells with functions to modify and contribute to the processing of rRNA, particularly during the synthesis of the ribosomal subunits [14][15][16][17]. The two main functions of snoRNA in relation to rRNA modifications involve methylation of the ribosomal subunits or 2′O-ribose-methylations and pseudouridylation to convert uridine into pseudouridine for the generation of mature rRNAs [18]. rRNAs and tRNAs are long and well-studied ncRNAs with known specific functions in mRNA translation. rRNAs are known to have a role in the assembly of ribosomal subunits and tRNAs in protein synthesis, particularly in the transfer of individual amino acid subunits into the ribosome during the translation of mRNA [19][20]. Notably, small RNA fragments derived from tRNAs can function as ncRNAs, as described below. The function of piRNAs is predominantly linked to transposons, particularly to protect the genome from invasive transposable elements in the germlines of animals through gene silencing [21]. This ncRNA is approximately 24–32 nucleotide bases and is transcribed from a series of repetitive elements within the genome known as piRNA clusters [22]. piRNAs interact with PIWI, a subfamily of ARGONAUTE proteins, to regulate their targets, including self-biogenesis [23]. In addition to the silencing of transposable elements, piRNAs also regulate DNA rearrangements, mRNA turnover and epigenetic programming [23][24].
2. MiRNA-Mediated Epigenetic Mechanisms
MiRNAs are a class of double-stranded RNAs (dsRNAs) that are approximately 22 nucleotide bases. This class of ncRNA is mostly responsible to silence mRNA translation by direct interaction with the transcript [25]. MiRNAs exert multiple cellular processes, including cell proliferation, adhesion, cell death and differentiation [26]. They were first characterized in Caenorhabditis elegans, with the discovery of lin-4, which was shown to silence lin-14 mRNA, which is a protein involved in the initial stage of larval (L1) development [27].
MiRNA gene transcription forms a stem-loop, double-stranded structure known as primary miRNA (pri-miRNA) in the nucleus (Figure 1) [28]. The pri-mRNA is processed, in the nucleus, by the RNase III enzyme Drosha [29] and its cofactor, Pasha, resulting in the formation of pri-miRNA into pre-mRNA. The pre-mRNA is further processed by the ATP-dependent protein/enzyme Dicer into a mature miRNA [30][31][32][33]. Once in the cytoplasm, one of the two strands of the mature miRNA is taken up by a member of the ARGONAUTE protein family. ARGONAUTE then delivers the miRNA to a target mRNA sequence, causing its degradation or preventing its translation [34]. ARGONAUTE forms complexes with different proteins in order to deliver the miRNA to its targets. For instance, the heat-shock protein 90 (Hsp90) can form a complex with the ARGONAUTE 1 (AGO1) in plants, along with the single miRNA strand [35]. This overall complex is known as the RNA-induced silencing complex or RISC. The single miRNA strand either binds with perfect or imperfect complementarity to the mRNA target, generally in the 3′ UTR, which leads to translation repression or mRNA degradation [36].
Figure 1. MicroRNA (MiRNA) biogenesis pathway. Transcription of the miRNA gene by RNA polymerase Pol II or Pol III produces a primary transcript (priRNA) that is cleaved by the ribonuclease III Drosha and processed by the double-stranded RNA-binding protein Pasha in the nucleus. This results in the formation of a precursor miRNA (pre-miRNA) hairpin, which is exported from the nucleus via exportin-5-mediated translocation. Once in the cytoplasm, the pre-miRNA is cleaved by the RNase DICER in a complex with the double-stranded RNA-binding protein TRBP to generate a miRNA duplex. The non-functional strand of the miRNA duplex is subjected to degradation, while the mature miRNA (functional strand) binds to ARGONAUTE proteins and the RNA-induced silencing complex (RISC). The mature miRNA guides RISC to silence mRNA targets by cleavage, translational repression or deadenylation.
SiRNAs are also small RNAs, commonly used experimentally as RNA interference or RNAi to silence genes. The use of RNAi was originally implemented in C. elegans in order to decrease the expression of specific genes and, since then, has been widely applied in varied experimental conditions [37]. This study found that double-stranded RNA interference (dsRNAi) molecules could be relatively more effective in silencing genes as compared to single-stranded siRNA. Shortly after this pioneering finding by Fire and colleagues in 1998, RNAi was implemented in the form of short hairpin RNAs (shRNAs) produced in plasmid vectors [38][39]. This followed the wide use of shRNAs through cell engineering with genomic-integrated shRNA [40]. Both siRNAs and shRNAs are used to silence genes for in vitro and in vivo experiments. In contrast to siRNAs, shRNA constructs are capable of DNA integration. After transcription, shRNAs are exported from the nucleus and recognized by Dicer in the cytosol to be processed into siRNA duplexes [41].
Post-transcriptional gene silencing (PTGS) and mRNA degradation allow miRNAs to regulate the epigenome through the downregulation of key epigenetic modifiers and to change the chromatin landscape [42]. Key examples of miRNA-interacting epigenetic factors include histone deacetylases (HDACs), histone methyltransferases (HMTs) and DNA methyltransferases (DNMTs) [42][43]. A list of miRNAs targeting specific repressive epigenetic modifiers is shown in Table 1. Thus, the upregulation of these miRNAs can specifically reactivate genes whose expressions were silenced by such epigenetic modifiers targeted by specific miRNAs. For instance, the miR-29 family has complementarities to the 3′ UTRs of DNMT3A and DNMT3B [44], resulting in their repression at the PTGS level. Similarly, miR-148 targets DNMT3B not at the 3′ UTRs but at the conserved protein-coding region [45]. Consequently, miR-148 interactions with DNMT3B mRNA lead to both PTGS, which could lead to translation deficiency or mRNA degradation [45]. DNA-methyl transferase 1 (DNMT1) is targeted by multiple miRNAs, including miR-148a, miR-152, miR-185 and miR-342. Many of the miRNAs listed in Table 1 have been reported to interact and repress the expression of the HMT, EZH2 (Enhancer of zeste homolog 2), which is part of the polycomb group repressive complex 2 (PRC2) [46]. EZH2 is targeted by several miRNAs, including miR-101, miR-137, miR-26a, miR-98, miR-124, miR-214 and let-7. In cases when miRNAs capable of suppressing DNMT1, DNMT3A, DNMT3B and EZH2 are decreased, it could lead to abnormal DNA methylation patterns to silence specific gene targets, resulting in cancer [47]. Abnormal DNA/histone methylation patterns could lead to the reactivation of oncogenes and/or the repression of tumor suppressors to facilitate cancer formation, progression and metastasis.
Table 1. MicroRNAs (MiRNAs) targeting epigenetic regulators. DNMT1: DNA-methyl transferase 1 and AML: acute myeloid leukemia.
|
miRNA |
Targets |
Function |
References |
|
miR-29 a, b, c |
DNMT3A and DNMT3B |
Tumor suppression by repression of de novo DNA methylation. Protects tumor-suppressor genes from been silenced by DNA methylation. |
Fabbri et al., 2007 [44] Suzuki et al., 2013 [47] |
|
miR-148 |
DNMT3B DNMT1 |
Negative feedback loop between DNMT1 and miR-148 in AML. Inhibition of cell proliferation and increase of apoptosis. |
Duursma et al., 2008 [45] Wang et al., 2019 [48] |
|
miR-449a |
HDAC1 |
Inhibition of tumor growth, invasion and metastasis. Promotes apoptosis and differentiation. |
Noonan et al., 2009 [49] Yong-Ming et al., 2017 [50] |
|
miR-152 miR-185 miR-342 |
DNMT1 |
DNA hypomethylation. Promotes the expression of tumor-suppressor genes. |
Suzuki et al., 2013 [47] |
|
miR-26a miR-98 miR-124 miR-214 let-7 miR-101 miR-137 |
EZH2 |
Prevents the progression of prostate cancer and metastasis. |
Suzuki et al., 2013 [47] |
In addition to miRNAs capable of regulating the epigenome, the expression of such miRNAs themselves can be regulated by epigenetic modifications. For instance, CpG islands, which are generally found at gene promoters, are also found in approximately half of all miRNA genes, which can consequently undergo aberrant DNA methylation and deregulated expression [42]. These modifications can lead to either the upregulation or downregulation of miRNA expressions that can be associated with different states of tumorigenesis.
References
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: From mechanism to therapy. Cell 2012, 150, 12–27.
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395.
- Verdin, E.; Ott, M. 50 years of protein acetylation: From gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. 2015, 16, 258–264.
- Feil, R.; Fraga, M.F. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2012, 13, 97–109.
- Tiffon, C. The impact of nutrition and environmental epigenetics on human health and disease. Int. J. Mol. Sci. 2018, 19, 3425.
- Wolffe, A.P.; Matzke, M.A. Epigenetics: Regulation through repression. Science 1999, 286, 481–486.
- Egger, G.; Liang, G.; Aparicio, A.; Jones, P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004, 429, 457–463.
- Chuang, J.C.; Jones, P.A. Epigenetics and microRNAs. Pediatr. Res. 2007, 61, 24–29.
- Kim, Y.J.; Maizel, A.; Chen, X. 2014. Traffic into silence: Endomembranes and post‐transcriptional RNA silencing. EMBO J. 2014, 33, 968–980.
- Tao, B.B.; Liu, X.Q.; Zhang, W.; Li, S.; Dong, D.; Xiao, M.; Zhong, J. Evidence for the association of chromatin and microRNA regulation in the human genome. Oncotarget 2017, 8, 70958–70966.
- Karijolich, J.; Yu, Y.T. Spliceosomal snRNA modifications and their function. RNA Biol. 2010, 7, 192–204.
- Jurica, M.S.; Moore, M.J. Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell 2003, 12, 5–14.
- Valadkhan, S. snRNAs as the catalysts of pre-mRNA splicing. Curr. Opin. Chem. Biol. 2005, 9, 603–608.
- Weinstein, L.B.; Steitz, J.A. Guided tours: From precursor snoRNA to functional snoRNP. Curr. Opin. Cell. Biol. 1999, 11, 378–384.
- Kiss, T. Small nucleolar RNA‐guided post‐transcriptional modification of cellular RNAs. EMBO J. 2001, 20, 3617–3622.
- Matera, A.G.; Terns, R.M.; Terns, M.P. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. 2007, 8, 209–220.
- Ono, M.; Yamada, K.; Avolio, F.; Scott, M.S.; van Koningsbruggen, S.; Barton, G.J.; Lamond, A.I. Analysis of human small nucleolar RNAs (snoRNA) and the development of snoRNA modulator of gene expression vectors. Mol. Biol. Cell 2010, 21, 1569–1584.
- Maden, B.E.H.; Hughes, J.M. Eukaryotic ribosomal RNA: The recent excitement in the nucleotide modification problem. Chromosoma 1997, 105, 391–400.
- Brimacombe, R.; Stiege, W. Structure and function of ribosomal RNA. Biochem. J. 1985, 229, 1–17.
- O’Donoghue, P.; Ling, J.; Söll, D. Transfer RNA function and evolution. RNA Biol. 2018, 15, 423–426.
- Ishizu, H.; Siomi, H.; Siomi, M.C. Biology of PIWI-interacting RNAs: New insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012, 26, 2361–2373.
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103.
- Ku, H.Y.; Lin, H. PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression. Natl. Sci. Rev. 2014, 1, 205–218.
- Liu, J.; Zhang, S.; Cheng, B. Epigenetic roles of PIWI interacting RNAs (piRNAs) in cancer metastasis. Oncol. Rep. 2018, 40, 2423–2434.
- Alberti, C.; Cochella, L. A framework for understanding the roles of miRNAs in animal development. Development 2017, 144, 2548–2559.
- Hwang, H.W.; Mendell, J.T. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer 2006, 94, 776–780.
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854.
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297.
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419.
- Nykänen, A.; Haley, B.; Zamore, P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 2001, 107, 309–321.
- Ketting, R.F.; Fischer, S.E.; Bernstein, E.; Sijen, T.; Hannon, G.J.; Plasterk, R.H. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001, 15, 2654–2659.
- Grishok, A.; Pasquinelli, A.E.; Conte, D.; Li, N.; Parrish, S.; Ha, I.; Baillie, D.L.; Fire, A.; Ruvkun, G.; Mello, C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 2001, 106, 23–34.
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366.
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduc. Target. Ther. 2016, 1, 1–9.
- Iki, T.; Yoshikawa, M.; Nishikiori, M.; Jaudal, M.C.; Matsumoto-Yokoyama, E.; Mitsuhara, I.; Meshi, T.; Ishikawa, M. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol. Cell 2010, 39, 282–291.
- Chen, C.Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86.
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811.
- Yu, J.Y.; DeRuiter, S.L.; Turner, D.L. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 2002, 99, 6047–6052.
- Miyagishi, M.; Taira, K. U6 promoter–driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 2002, 20, 497–500.
- Abbas-Terki, T.; Blanco-Bose; W.; Déglon, N.; Pralong, W.; Aebischer, P. Lentiviral-mediated RNA interference. Hum. Gene Ther. 2002, 13, 2197–2201.
- Moore, C.B.; Guthrie, E.H.; Huang, M.T.; Taxman, D.J.; Short hairpin RNA (shRNA): Design, delivery, and assessment of gene knockdown. Methods Mol Biol 2010, 629, 141-158.
- Bianchi, M.; Renzini, A.; Adamo, S.; Moresi, V. Coordinated actions of microRNAs with other epigenetic factors regulate skeletal muscle development and adaptation. Int. J. Mol. Sci. 2017, 18, 840.
- Grunstein, M. Histone acetylation in chromatin structure and transcription. Nature 1997, 389, 349–352.
- Fabbri, M.; Garzon, R.; Cimmino, A.; Liu, Z.; Zanesi, N.; Callegari, E.; Liu, S.; Alder, H.; Costinean, S.; Fernandez-Cymering, C.; et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA 2007, 104, 15805–15810.
- Duursma, A.M.; Kedde, M.; Schrier, M.; Le Sage, C.; Agami, R. miR-148 targets human DNMT3b protein coding region. RNA 2008, 14, 872–877.
- Margueron, R.; Li, G.; Sarma, K.; Blais, A.; Zavadil, J.; Woodcock, C.L.; Dynlacht, B.D.; Reinberg, D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 2008, 32, 503–518.
- Suzuki, H.; Maruyama, R.; Yamamoto, E.; Kai, M. Epigenetic alteration and microRNA dysregulation in cancer. Front. Genet. 2013, 4, 258.
- Wang, X.X.; Zhang, H.; Li, Y. Preliminary study on the role of miR 148a and DNMT1 in the pathogenesis of acute myeloid leukemia. Mol. Med. Rep. 2019, 4, 2943–2952.
- Noonan, E.;J.; Place, R.F.; Pookot, D.; Basak, S.; Whitson, J.M.; Hirata, H.; Giardina, C.; Dahiya, R. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene 2009, 28, 1714–1724.
- Yong-Ming, H.; Ai-Jun, J.; Xiao-Yue, X.; Jian-Wei, L.; Chen, Y.; Ye, C. miR-449: A potential therapeutic agent for cancer. Anticancer Drugs 2017, 28, 1067–1078.