
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Stéphane Maury | + 1697 word(s) | 1697 | 2020-09-16 08:22:33 | | | |
| 2 | Vivi Li | -101 word(s) | 1596 | 2020-09-21 06:05:43 | | |
Video Upload Options
Epigenetics refers to a scientific domain studying all the processes affecting the expression of genes and/or the activity of transposable elements (TEs) without altering the DNA sequence that may be heritable by mitosis (during development) and/or meiosis (across generations). Forest trees are sessile, perennial, and modular organisms with complex life cycles that are often challenged by environmental variations such as actual climate changes during their long-lifespan. Surviving tree populations can respond to these environmental changes through complex and interacting mechanisms and notably using epigenetics.
1. Epigenetics in Plants
Epigenetics has been initially defined as “the branch of biology which studies the causal interactions between genes and their products, which bring the phenotype into being” [1]. Here, we define it as the study of all the processes affecting the expression of genes and/or the activity of transposable elements (TEs) without altering the DNA sequence that may be heritable by mitosis (during development) and/or meiosis (across generations) [2][3][4] (Figure 1).
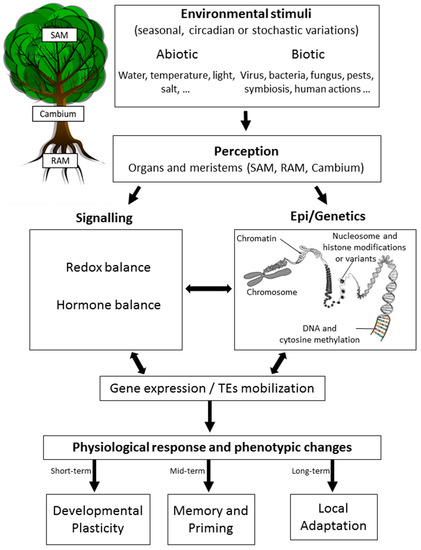
Figure 1. Epigenetic response to environmental changes in trees. Trees are known to recognize various abiotic and/or biotic stimuli occurring rhythmically (circadian or seasonal) or stochastically. These changes are perceived at different tissue levels, with most studies focusing on the leaves, roots, and meristems (SAM, shoot apical meristem; Cambium; or RAM, root apical meristem). Perception is then followed by signaling mechanisms that may include changes in the redox or hormonal balance, which have been shown to be related to epigenetic changes (chromatin remodeling, DNA methylation, non-coding RNA mechanisms (not shown), and histone modifications and variants). This complex crosstalk between signaling processes, epigenetics, and genetics results in an altered gene expression status and/or the mobilization of transposable elements (TEs). A physiological response is then observed together with phenotypic changes that allow trees to acclimate to the environmental changes initially sensed depending on the time-scale considered and the heritable transmission of epigenetic changes.
2. Promises of Epigenetics for Tree Improvement, Breeding, Conservation of Genetic Resources and Forest Management
The potential of epigenetics in plant breeding is already being considered in crops [5][6][7][8][9][10]. Only limited studies have been implemented with trees, and results coming from crops will need to be adapted to fit with the specific features of tree breeding and forest management [11]. Although most of the studies on forest trees have focused on the role of epigenetics in tree development, and response to environmental changes and priming (Table S1), recent studies have pointed out the potential relevance of epigenetics in tree improvement. Three main approaches using epigenetics may be foreseen: (i) Exploit natural or artificially-induced epigenetic diversity; (ii) use epigenetic marks in addition to classic genetic markers for trait improvement with trans-omics approach [12], statistics and modeling, and (iii) use epigenome editing [13]. Epigenetics could be used to expand the material for tree breeders that may be used by sexual or asexual propagation methods.
The best-known example in trees for exploiting natural or induced epigenetic diversity is the existence of an epigenetic memory mechanism that operates during embryo development in Norway spruce (see above section). This process adjusts the timing of bud burst in the progeny, but also in genetically identical epitypes in a manner usually associated with ecotypes in accordance with the temperature conditions during embryogenesis [14][15][16][17][18]. Kvaalen and Johnsen (2008) [16] demonstrated that environmentally induced epigenetic memory during somatic embryogenesis can give similar results for phenology as to those produced by provenance separation of 4–6° latitude. Environmentally induced epigenetic memory during somatic embryogenesis could potentially be used in tree breeding to prime trees regarding their phenology for their first years of plantation along a latitudinal gradient. This promising application opens up many possibilities for forest tree research considering the increasing literature on tree priming (Table S1), and unravels the importance of epigenetic natural diversity that should be taken into consideration for forest breeding, conservation of genetic resources and forest management and protection. Another example of exploiting natural epigenetic variation is emerging from the role of epigenetics in tree heterosis that has been evaluated in Populus deltoides [19]. Findings indicate that methylation patterns of the two P. deltoides parental lines are both partially and dynamically passed on to their intraspecific hybrids, resulting in a non-additive methylation pattern in F1 hybrids. However, studies on epigenetic and heterosis are still necessary for trees and will have to be evaluated by tree breeders.
Another perspective of forest tree epigenetics comes from studies conducted on Arabidopsis and crops concerning sRNAs and cross-kingdom RNA interference (RNAi) [20]. Cai et al. (2018) [21] have shown that Arabidopsis has adapted its exosome-mediated cross-kingdom RNAi as part of its immune responses against pathogens. Pathogens and pests can thus be controlled by sRNAs, targeting their essential or pathogenicity genes, raising the possibility of plants to be protected from diseases by a novel eco-friendly, durable, and highly specific RNA fungicide or pesticide [22][23]. In addition, there is growing evidence to indicate that epigenetic mechanisms directly participate in plant immune responses. However, the evidence of transgenerational inheritance of pathogen-induced defense priming is still a matter of debate [24]. Studies have shown that biotic stresses can also trigger an increase in the overall level of genomic methylation. Curiously, the methylation levels of some pathogen responses or resistance genes are reduced [25]. We still need to understand exactly how epigenetics controls trees’ defenses against disease to then translate this knowledge into practical actions. By exploring natural and induced resistance, we may implement new breeding strategies for preventing disease while reducing the global reliance on harmful pesticides, a field of research still in its infancy for trees.
The application of epigenetic markers to tree breeding has been tested over the last few years, including studies highlighting the extent and variation of genome-wide DNA methylation in natural populations of trees. Ci et al. (2015) [26] investigated the variation in DNA methylation and whether this variation correlates with important plant traits, including leaf shape and photosynthesis in Populus simonii, indicating that epigenetics bridges environmental and genetic factors in affecting plant growth and development. Sow et al. (2018) [27] reported the first estimate of narrow-sense heritability (h2) and phenotypic differentiation (Pst) for global DNA methylation in trees by assessing global DNA methylation variations in Populus nigra clones from natural populations under varying soil water availability. Regardless of water regimes, values of h2 and Pst were comparable to those found for shoot biomass production, a known heritable trait in poplar. Therefore, global DNA methylation, being genetically and environmentally determined in these populations, could be used as a potential marker for population differentiation, performance, and selection under stressful conditions. To explore the epigenetic diversity in breeding programs, we need to expand our knowledge regarding the link between DNA methylation and economic traits in forest trees. Ma et al. (2012) [28] investigated 130 intraspecific hybrids of Populus tomentosa and concluded that the regions defined by the MSAP candidate markers are linked to genes that are essential for photosynthetic traits that respond to DNA methylation which affect growth traits.
The potential of epigenetic markers in quantitative breeding approaches has been recently suggested by Baison et al. (2019) [29]. The authors identified 52 QTLs (Quantitative Trait Loci) of wood properties in Norway spruce for marker-assisted breeding. However, these QTLs explain a small proportion of the genetic variation, in line with previous studies examining genetic variation in complex traits in coniferous species using forward genetic approaches. They suggest that this could be due to several factors, including epigenetic variation, highlighting the need for sophisticated epi/genotyping tools, as well as a combination of advanced statistical models, such as regional heritability mapping. Recently, Champigny et al. (2019) [30] applied statistical learning experiments to genetically diverse populations of Populus balsamifera trees grown at two common garden sites and showed that traits in novel genotypes can be modelled using small numbers of methylated DNA predictors. Indeed, significant phenotypic variances in quantitative traits of the wood were explained by the natural variation of DNA methylation, such as total biomass (57.5%), wood density (40.9%), soluble lignin (25.3%), and carbohydrate content in the cell wall (mannose: 44.8%). The authors proposed that DNA methylation-based models can be used as a strategy to validate the identity, provenance, or quality of agroforestry products. While this approach presents innovative perspectives for forest trees, it should be evaluated in terms of cost, technical, and analytical efforts.
The last approach to use epigenetics for tree improvement is epigenome editing, i.e., a type of genetic engineering in which the epigenome is modified at specific sites using engineered molecules targeted to those sites. Genome editing in trees has been recently reviewed by Bewg et al. (2018) [13], suggesting a great potential for stable CRISPR-induced mutations and associated phenotypes over multiple clonal generations. Authors suggest that this technology can be used for the commercial production of elite trees that relies on vegetative propagation. The potential of epigenome editing to control recombination in plant breeding has been recently reviewed [31]. Manipulating the rate and positions of crossovers to increase the genetic variation accessible to breeders is of main interest. Epi/genome editing at desired sites of recombination, and manipulation of crossovers factors, are applicable approaches for achieving this goal, reducing the time and expense associated with traditional breeding, revealing inaccessible genetic diversity, and increasing control over the inheritance of preferred haplotypes [31]. However, there is no study on epigenome editing in trees, and the feasibility of such technological advances, depending on use regulations in different countries, will have to be evaluated by tree breeders in the future.
Supplementary Materials: The following are available online at https://www.mdpi.com/1999-4907/11/9/976/s1. Table S1: Current state of epigenetics in trees for development, abiotic stress or priming, biotic stress or priming, and markers, breeding and biotechnology topics.
References
- C. H. Waddington; CANALIZATION OF DEVELOPMENT AND THE INHERITANCE OF ACQUIRED CHARACTERS. Nature 1942, 150, 563-565, 10.1038/150563a0.
- Bossdorf, O.; Richards, C.L.; Pigliucci, M. Epigenetics for ecologists. Ecol. Lett. 2007, 11, 106–115.
- Richards, E.J. Natural epigenetic variation in plant species: A view from the field. Curr. Opin. Plant Biol. 2011, 14, 204–209.
- Ledón-Rettig, C.C. Ecological Epigenetics: An Introduction to the Symposium. Integr. Comp. Biol. 2013, 53, 307–318.
- Christina L. Richards; Conchita Alonso; Claude Becker; Oliver Bossdorf; Etienne Bucher; Maria Colome-Tatche; Walter Durka; Jan Engelhardt; Bence Gáspár; Andreas Gogol-Döring; et al.Ivo GrosseThomas P. Van GurpKatrin HeerIlkka KronholmChristian LampeiVít LatzelMarie MirouzeLars OpgenoorthOvidiu PaunSonja J. ProhaskaStefan A. RensingPeter F. StadlerEmiliano TrucchiKristian K. UllrichKoen J. F. Verhoeven Ecological plant epigenetics: Evidence from model and non-model species, and the way forward. Ecology Letters 2017, 20, 1576-1590, 10.1111/ele.12858.
- Gallusci, P.; Dai, Z.; Génard, M.; Gauffretau, A.; Leblanc-Fournier, N.; Richard-Molard, C.; Vile, D.; Brunel-Muguet, S. Epigenetics for Plant Improvement: Current Knowledge and Modeling Avenues. Trends Plant Sci. 2017, 22, 610–623.
- Springer, N.M.; Schmitz, R.J. Exploiting induced and natural epigenetic variation for crop improvement. Nat. Rev. Genet. 2017, 18, 563–575.
- Achour, Z.; Archipiano, M.; Barneche, F.; Baurens, C.; Beckert, M.; Ben, C.; Epigenetics in Plant Breeding. Article de Positionnement du GROUPEMENT D’intérêt Scientifique Biotechnologies Vertes et de L’alliance Nationale de Recherche Pour L’environnement. 2017. Available online: http://www.gisbiotechnologiesvertes.com/fr/publications/position-paperepigenetics-in-plant-breeding (accessed on 13 February 2017).
- Gallusci, P.; Bucher, E.; Mirouze, M. Preface. In Advances in Botanical Research; Mirouze, M., Bucher, E., Gallusci, P., Eds.; Plant Epigenetics Coming of Age for Breeding Applications; Academic Press: Cambridge, MA, USA, 2018; Volume 88, pp. 15–18.
- Ryder, P.; McKeown, P.C.; Fort, A.; Spillane, C. Epigenetics and Heterosis in Crop Plants. In Epigenetics in Plants of Agronomic Importance: Fundamentals and Applications; Alvarez-Venegas, R., De-la-Peña, C., Casas-Mollano, J.A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 129–147.
- Sow, M.D.; Allona, I.; Ambroise, C.; Conde, D.; Fichot, R.; Gribkova, S.; Jorge, V.; Le-Provost, G.; Pâques, L.; Plomion, C.; et al. Epigenetics in Forest Trees. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2018; Volume 88, pp. 387–453.
- Katsuyuki Yugi; Hiroyuki Kubota; Atsushi Hatano; Shinya Kuroda; Trans-Omics: How To Reconstruct Biochemical Networks Across Multiple ‘Omic’ Layers. Trends in Biotechnology 2016, 34, 276-290, 10.1016/j.tibtech.2015.12.013.
- William Patrick Bewg; Dong Ci; Chung-Jui Tsai; Genome Editing in Trees: From Multiple Repair Pathways to Long-Term Stability. Frontiers in Plant Science 2018, 9, 9, 10.3389/fpls.2018.01732.
- Igor A. Yakovlev; Daniel K.A. Asante; Carl G. Fossdal; Olavi Junttila; Oystein Johnsen; Differential gene expression related to an epigenetic memory affecting climatic adaptation in Norway spruce. Plant Science 2011, 180, 132-139, 10.1016/j.plantsci.2010.07.004.
- Johnsen, O.; Daehlen, O.G.; Østreng, G.; Skrøppa, T.; Dæhlen, O.G. Daylength and temperature during seed production interactively affect adaptive performance of Picea abies progenies. New Phytol. 2005, 168, 589–596.
- Kvaalen, H.; Øystein, J. Timing of bud set in Picea abies is regulated by a memory of temperature during zygotic and somatic embryogenesis. New Phytol. 2007, 177, 49–59.
- Carneros, E.; Yakovlev, I.; Viejo, M.; Olsen, J.E.; Fossdal, C.G. The epigenetic memory of temperature during embryogenesis modifies the expression of bud burst-related genes in Norway spruce epitypes. Planta 2017, 246, 553–566.
- Tore Skrøppa; Mari Mette Tollefsrud; Christoph Sperisen; Øystein Johnsen; Rapid change in adaptive performance from one generation to the next in Picea abies—Central European trees in a Nordic environment. Tree Genetics & Genomes 2009, 6, 93-99, 10.1007/s11295-009-0231-z.
- Ming Gao; Qinjun Huang; Yanguang Chu; Changjun Ding; Bingyu Zhang; Xiaohua Su; Analysis of the leaf methylomes of parents and their hybrids provides new insight into hybrid vigor in Populus deltoides. BMC Genetics 2014, 15, S8-S8, 10.1186/1471-2156-15-S1-S8.
- Arne Weiberg; Marschal Bellinger; Hailing Jin; Conversations between kingdoms: small RNAs. Current Opinion in Biotechnology 2015, 32, 207-215, 10.1016/j.copbio.2014.12.025.
- Jian-Min Zhou; Xiangxiu Liang; Cai Q; He B; Kogel Kh; Jin H; Faculty Opinions recommendation of Cross-kingdom RNA trafficking and environmental RNAi-nature's blueprint for modern crop protection strategies.. Faculty Opinions – Post-Publication Peer Review of the Biomedical Literature 2018, 46, 58-64, 10.3410/f.732871062.793551297.
- Cai, Q.; He, B.; Kogel, K.H.; Jin, H. Cross-kingdom RNA trafficking and environmental RNAi—Nature’s blueprint for modern crop protection strategies. Curr. Opin. Microbiol. 2018, 46, 58–64.
- Muhammad, T.; Zhang, F.; Zhang, Y.; Liang, Y. RNA Interference: A Natural Immune System of Plants to Counteract Biotic Stressors. Cells 2019, 8, 38.
- Juan S. Ramirez-Prado; Aala A Abulfaraj; Naganand Rayapuram; Moussa Benhamed; Heribert Hirt; Plant Immunity: From Signaling to Epigenetic Control of Defense. Trends in Plant Science 2018, 23, 833-844, 10.1016/j.tplants.2018.06.004.
- Hai Peng; Jing Zhang; Plant genomic DNA methylation in response to stresses: Potential applications and challenges in plant breeding. Progress in Natural Science: Materials International 2009, 19, 1037-1045, 10.1016/j.pnsc.2008.10.014.
- Dong Ci; Yuepeng Song; Min Tian; Deqiang Zhang; Methylation of miRNA genes in the response to temperature stress in Populus simonii. Frontiers in Plant Science 2015, 6, 921, 10.3389/fpls.2015.00921.
- Mamadou Dia Sow; Vincent Segura; Sylvain Chamaillard; Véronique Jorge; Alain Delaunay; Clément Lafon Placette; Régis Fichot; Patricia Faivre-Rampant; Marc Villar; Franck Brignolas; et al.Stéphane Maury Narrow-sense heritability and PST estimates of DNA methylation in three Populus nigra L. populations under contrasting water availability. Tree Genetics & Genomes 2018, 14, 78, 10.1007/s11295-018-1293-6.
- Kaifeng Ma; Yuepeng Song; Xibing Jiang; Zhiyi Zhang; Bailian Li; Deqiang Zhang; Photosynthetic response to genome methylation affects the growth of Chinese white poplar. Tree Genetics & Genomes 2012, 8, 1407-1421, 10.1007/s11295-012-0527-2.
- John Baison; Amaryllis Vidalis; Linghua Zhou; Zhi‐Qiang Chen; Zitong Li; Mikko J. Sillanpää; Carolina Bernhardsson; Douglas Scofield; Nils Forsberg; Thomas Grahn; et al.Lars OlssonBo KarlssonHarry WuPär K. IngvarssonSven‐Olof LundqvistTotte NiittyläM Rosario García‐Gil Genome‐wide association study identified novel candidate loci affecting wood formation in Norway spruce. The Plant Journal 2019, 100, 83-100, 10.1111/tpj.14429.
- Marc J. Champigny; Faride Unda; Oleksandr Skyba; Raju Y. Soolanayakanahally; Shawn D. Mansfield; M. M. Campbell; Learning from methylomes: epigenomic correlates of Populus balsamifera traits based on deep learning models of natural DNA methylation. Plant Biotechnology Journal 2019, 18, 1361-1375, 10.1111/pbi.13299.
- Ella Taagen; Adam J. Bogdanove; Mark E. Sorrells; Counting on Crossovers: Controlled Recombination for Plant Breeding. Trends in Plant Science 2020, 25, 455-465, 10.1016/j.tplants.2019.12.017.




