
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Emma Poole | + 871 word(s) | 871 | 2020-07-27 08:53:37 | | | |
| 2 | Felix Wu | -54 word(s) | 817 | 2020-11-21 14:02:05 | | | | |
| 3 | Felix Wu | -54 word(s) | 817 | 2020-11-21 14:02:27 | | |
Video Upload Options
Human cytomegalovirus (HCMV) establishes both latent (where the virus is maintained in an absence of the production of infectious virions) and lytic (during primary infection and reactivation) phases of infection. The study of latency has been historically more difficult to study than that of lytic infection due to the relative quiescence of the virus in host cells and the limited cell types that support HCMV latency. With the advent of engineered HCMV viruses which express fluorescent tags that enable latently infected cells to be detected and isolated, it has been possible to analyse latently infected cells in much greater detail. In this article, we review studies which have used such systems to analyse and interrogate the latency associated proteome. These have led to the identification of cellular proteins and pathways important for latency and, consequently, potential therapeutic intervention drugs based on a greater understanding of latent infection.
1. Introduction
Human cytomegalovirus (HCMV) is a species-specific pathogen which is carried by approximately 60–80% of the population, depending on demographics [1]. Front-line anti-virals for HCMV are limited due to poor bioavailability and drug resistance. Additionally, currently available drugs target the lytic phase of infection rather than the latent phase (the phases of the lifecycle are described in detail below) [2][3]. Therefore, the need to target latently infected cells could be crucial, particularly in settings such as solid organ or stem cell transplantation where latent virus in the graft itself can be transferred to an immune-suppressed donor or a seropositive graft recipient’s own virus can reactivate.
2. HCMV Lytic and Latent Lifecycle
HCMV is a large double-stranded DNA virus with a protracted lytic lifecycle; up to 72 h in some cell types. This commences with immediate early (IE) gene expression, then early (E) gene expression and finally, late gene expression (L) [4]. The lytic lifecycle can take place in a wide range of terminally differentiated cell types, including fibroblasts, macrophages’ endothelial cells, resulting in full productive virus replication. The ability of the virus to establish a lifelong infection in the host is, at least in part, due to the ability of the virus to also be able to establish a latent infection during which time viral genome is carried in cells but IE gene expression is suppressed and infectious virions are not produced [5]. One established site of HCMV latency is in the cells of the myeloid lineage [5] where, in vivo, virus is carried in myeloid progenitors such as CD34+ progenitor cells and their derivative CD14+ monocytes. Then, as the cells differentiate along the myeloid lineage to terminally differentiated cell types such as macrophages or dendritic cells, the virus reactivates leading to the initiation of IE gene expression, the full temporal lytic gene cascade and ultimately, progeny virions [5].
3. Studying the Proteome
During lytic infection, temporal changes in the viral proteome [4] as well as changes in the cell proteome mediated by lytic infection have been extensively analysed [4][6]. Such studies have been expanded upon, using for instance shRNA or CRISP-R libraries, to identify mechanisms by which viral-viral or viral-cellular protein–protein interactions regulate both cellular and viral gene expression during lytic infection [4][7]. Such high-throughput approaches to understanding HCMV lytic infection have been recently reviewed [7][8][9]. In this review, we discuss total proteomic analyses that have been carried out to interrogate cellular changes during HCMV latency.
4. Proteome Changes Induced by Expression of Latency-Associated Proteins in Isolation
Though unbiased proteomic screens are a powerful way to interrogate latency-associated changes in cellular proteins, some targets could possibly be missed, especially because viral latency-associated gene expression is known to be very low in the latently infected cells. One way to address this is to overexpress putative latency associated genes in isolation. Another advantage of this targeted approach is that a screen which identifies changes in total cellular proteins induced by latency will not, inherently, identify which viral gene product has caused these changes. Therefore, a number of studies have analysed changes in myeloid cells which over-express specific latency-associated proteins. To date, this has been carried out for two viral genes known to be expressed during latency by overexpressing them in THP1 myelomonocytic cells, a cell type known to support HCMV latency and reactivation [10][11][12]. In this entry, we have concentrated on changes to the cellular proteome. Many of these changes (summarised in Figure 1) have identified mechanisms by which latent infection modifies, transendothelial migration and avoidance of neutrophil killing.
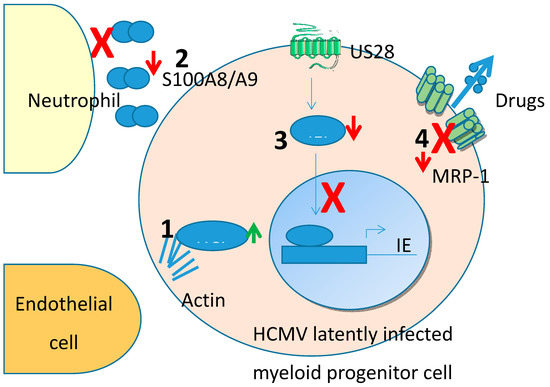
Figure 1. Cellular protein changes identified from unbiased proteomic screens in Human cytomegalovirus (HCMV) latency. (1) HCLS1 is upregulated which regulates actin stabilisation and, therefore, cell motility leading to enhanced endothelial cell attachment and transit. (2) S100A8/A9 are downregulated preventing the chemoattraction of neutrophils which could otherwise target and kill latently infected cells. (3) IFI16 is downregulated by latency-associated protein US28 and prevents NFkB activation of the MIEP and therefore helps aid the maintenance of latency. (4) Drug transporter MRP-1 is downregulated during latency by latency associated protein UL138 which renders cells sensitive to drug-mediated killing.
References
- Cannon, M.J.; Schmid, D.S.; Hyde, T.B. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev. Med. Virol. 2010, 20, 202–213.
- Ahmed, A. Antiviral treatment of cytomegalovirus infection. Infect. Disord. Drug Targets 2010, 11, 475–503.
- Ehlert, K.; Groll, A.H.; Kuehn, J.; Vormoor, J. Treatment of refractory CMV-infection following hematopoietic stem cell transplantation with the combination of foscarnet and leflunomide. Klin. Padiatr. 2006, 218, 180–184.
- Michael P. Weekes; Peter Tomasec; Edward L. Huttlin; Ceri A. Fielding; David Nusinow; Richard J. Stanton; Eddie C.Y. Wang; Rebecca Aicheler; Isa Murrell; Gavin W.G. Wilkinson; et al.Paul J. LehnerSteven P. Gygi Quantitative Temporal Viromics: An Approach to Investigate Host-Pathogen Interaction. Cell 2014, 157, 1460-1472, 10.1016/j.cell.2014.04.028.
- John Sinclair; Emma Poole; Human cytomegalovirus latency and reactivation in and beyond the myeloid lineage. Future Virology 2014, 9, 557-563, 10.2217/fvl.14.34.
- Dumortier, J.; Streblow, D.N.; Moses, A.V.; Jacobs, J.M.; Kreklywich, C.N.; Camp, D.; Smith, R.D.; Orloff, S.L.; Nelson, J.A. Human cytomegalovirus secretome contains factors that induce angiogenesis and wound healing. J. Virol. 2008, 82, 6524–6535.
- Lee, C.H.; Grey, F. Systems Virology and Human Cytomegalovirus: Using High Throughput Approaches to Identify Novel Host-Virus Interactions During Lytic Infection. Front. Cell Infect Microbiol. 2020, 10, 280.
- Arend, K.C.; Lenarcic, E.M.; Vincent, H.A.; Rashid, N.; Lazear, E.; McDonald, I.M.; Gilbert, T.S.; East, M.P.; Herring, L.E.; Johnson, G.L.; et al. Kinome Profiling Identifies Druggable Targets for Novel Human Cytomegalovirus (HCMV) Antivirals. Mol. Cell Proteom. 2017, 16 (Suppl. 1), S263–S276.
- Cohen, Y.; Stern-Ginossar, N. Manipulation of host pathways by human cytomegalovirus: Insights from genome-wide studies. Semin. Immunopathol. 2014, 36, 651–658.
- Patrick S. Beisser; Lysiane Laurent; Jean-Louis Virelizier; Susan Michelson; Human Cytomegalovirus Chemokine Receptor Gene US28 Is Transcribed in Latently Infected THP-1 Monocytes. Journal of Virology 2001, 75, 5949-5957, 10.1128/jvi.75.13.5949-5957.2001.
- Lau, B.; Poole, E.; Van Damme, E.; Bunkens, L.; Sowash, M.; King, H.; Murphy, E.; Wills, M.; Van Loock, M.; Sinclair, J. Human cytomegalovirus miR-UL112-1 promotes the down-regulation of viral immediate early-gene expression during latency to prevent T-cell recognition of latently infected cells. J. Gen. Virol. 2016, 97, 2387–2398.
- Poole, E.L.; Kew, V.G.; Lau, J.C.H.; Murray, M.J.; Stamminger, T.; Sinclair, J.H.; Reeves, M.B. A Virally Encoded DeSUMOylase Activity Is Required for Cytomegalovirus Reactivation from Latency. Cell Rep. 2018, 24, 594–606.




