
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Hakim Manghwar | + 1908 word(s) | 1908 | 2022-01-17 10:31:07 | | | |
| 2 | Yvaine Wei | Meta information modification | 1908 | 2022-01-18 06:44:41 | | | | |
| 3 | Hakim Manghwar | + 152 word(s) | 2060 | 2022-06-10 09:45:07 | | |
Video Upload Options
The endoplasmic reticulum (ER) is an important subcellular organelle, primarily recognized as a checkpoint for protein folding. It plays an essential role in ensuring the proper folding and maturation of newly secreted and transmembrane proteins. Different processes are activated when around one-third of newly synthesized proteins enter the ER in the eukaryote cells, such as glycosylation, folding, and/or the assembling of these proteins into protein complexes. However, protein folding in the ER is an error-prone process whereby various stresses easily interfere, leading to the accumulation of unfolded/misfolded proteins and causing ER stress. The unfolded protein response (UPR) is a process that involves sensing ER stress. Many strategies have been developed to reduce ER stress, such as UPR, ER-associated degradation (ERAD), and autophagy.
1. Introduction
2. Endoplasmic Reticulum (ER)
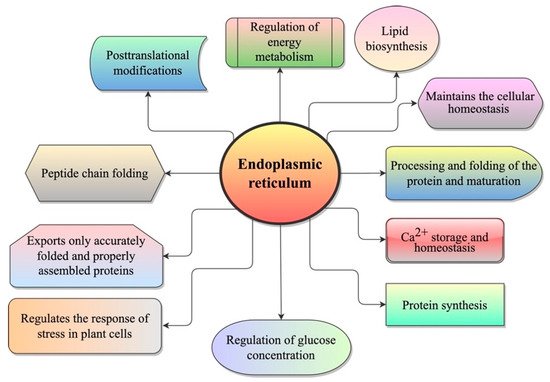
3. ER Stress
4. Chemical Inducers for the Accumulation of the Unfolded Protein
4.1. Tunicamycin (TM) Stress
4.2. Dithiothreitol (DTT) Stress
5. UPR Signaling in Plant Development
6. UPR Signaling in Different Stresses
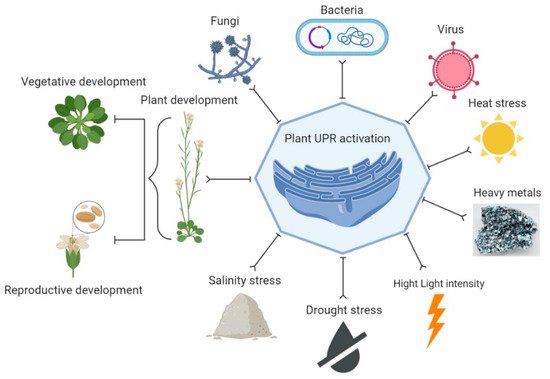
7. Strategies to Reduce ER Stress
7.1. Unfolded Protein Response (UPR)
7.2. Mechanism of UPR Signaling Pathway in Plants
7.2.1. Regulated IRE-1 Dependent Splicing (RIDS)
7.2.2. ER-Associated Degradation (ERAD)
7.2.3. Autophagy
References
- Millar, A.J. The intracellular dynamics of circadian clocks reach for the light of ecology and evolution. Ann. Rev. Plant Biol. 2016, 67, 595–618.
- de Souza, A.; Wang, J.-Z.; Dehesh, K. Retrograde signals: Integrators of interorganellar communication and orchestrators of plant development. Ann. Rev. Plant Biol. 2017, 68, 85–108.
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88–95.
- Su, Z.; Ma, X.; Guo, H.; Sukiran, N.L.; Guo, B.; Assmann, S.M.; Ma, H. Flower development under drought stress: Morphological and transcriptomic analyses reveal acute responses and long-term acclimation in Arabidopsis. Plant Cell 2013, 25, 3785–3807.
- Hakim; Ullah, A.; Hussain, A.; Shaban, M.; Khan, A.H.; Alariqi, M.; Gul, S.; Jun, Z.; Lin, S.; Li, J.; et al. Osmotin: A plant defense tool against biotic and abiotic stresses. Plant Physiol. Biochem. 2018, 123, 149–159.
- Liu, L.; Li, J. Communications Between the Endoplasmic Reticulum and Other Organelles During Abiotic Stress Response in Plants. Front. Plant Sci. 2019, 10, 749.
- Schuldiner, M.; Schwappach, B. From rags to riches—The history of the endoplasmic reticulum. Biochim. Biophys. Acta. Mol. Cell Res. 2013, 1833, 2389–2391.
- Westrate, L.; Lee, J.; Prinz, W.; Voeltz, G. Form follows function: The importance of endoplasmic reticulum shape. Ann. Rev. Biochem. 2015, 84, 791–811.
- Stefano, G.; Brandizzi, F. Advances in plant ER architecture and dynamics. Plant Physiol. 2018, 176, 178–186.
- Liu, Z.; Lv, Y.; Zhao, N.; Guan, G.; Wang, J. Protein kinase R-like ER kinase and its role in endoplasmic reticulum stress-decided cell fate. Cell Death Dis. 2015, 6, e1822.
- Schwarz, D.S.; Blower, M.D. The endoplasmic reticulum: Structure, function and response to cellular signaling. Cell Mol. Life Sci. 2016, 73, 79–94.
- Benham, A.M. Endoplasmic Reticulum redox pathways: In sickness and in health. FEBS J. 2019, 286, 311–321.
- Schröder, M.; Kaufman, R.J. The mammalian unfolded protein response. Ann. Rev. Biochem. 2005, 74, 739–789.
- Bao, Y.; Howell, S.H. The unfolded protein response supports plant development and defense as well as responses to abiotic stress. Front. Plant Sci. 2017, 8, 344.
- Nawkar, G.M.; Lee, E.S.; Shelake, R.M.; Park, J.H.; Ryu, S.W.; Kang, C.H.; Lee, S.Y. Activation of the transducers of unfolded protein response in plants. Front. Plant Sci. 2018, 9, 214.
- Merksamer, P.I.; Trusina, A.; Papa, F.R. Real-time redox measurements during endoplasmic reticulum stress reveal interlinked protein folding functions. Cell 2008, 135, 933–947.
- Merksamer, P.I.; Papa, F.R. The UPR and cell fate at a glance. J. Cell Sci. 2010, 123, 1003–1006.
- Kabir, M.F.; Kim, H.-R.; Chae, H.-J. Endoplasmic Reticulum Stress and Autophagy. In Endoplasmic Reticulum; IntechOpen: London, UK, 2018.
- Chen, Q.; Yu, F.; Xie, Q. Insights into endoplasmic reticulum-associated degradation in plants. New Phytol. 2020, 226, 345–350.
- Schubert, U.; Anton, L.C.; Gibbs, J.; Norbury, C.C.; Yewdell, J.W.; Bennink, J.R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 2000, 404, 770–774.
- Hetz, C.; Chevet, E.; Oakes, S.A. Proteostasis control by the unfolded protein response. Nat. Cell Biol. 2015, 17, 829–838.
- Kim, S.; Choi, Y.; Kwon, C.; Yun, H.S. Endoplasmic reticulum stress-induced accumulation of VAMP721/722 requires CALRETICULIN 1 and CALRETICULIN 2 in Arabidopsis. J. Integr. Plant Biol. 2019, 61, 974–980.
- Ruberti, C.; Kim, S.-J.; Stefano, G.; Brandizzi, F. Unfolded protein response in plants: One master, many questions. Curr. Opin. Plant Biol. 2015, 27, 59–66.
- Park, C.-J.; Park, J.M. Endoplasmic reticulum plays a critical role in integrating signals generated by both biotic and abiotic stress in plants. Front. Plant Sci. 2019, 10, 399.
- Martínez, I.M.; Chrispeels, M.J. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 2003, 15, 561–576.
- Carvalho, H.H.; Brustolini, O.J.; Pimenta, M.R.; Mendes, G.C.; Gouveia, B.C.; Silva, P.A.; Silva, J.C.F.; Mota, C.S.; Soares-Ramos, J.R.; Fontes, E.P. The molecular chaperone binding protein BiP prevents leaf dehydration-induced cellular homeostasis disruption. PLoS ONE 2014, 9, e86661.
- Hauptmann, P.; Riel, C.; Kunz-Schughart, L.A.; Fröhlich, K.U.; Madeo, F.; Lehle, L. Defects in N-glycosylation induce apoptosis in yeast. Mol. Microbiol. 2006, 59, 765–778.
- Iwata, Y.; Nishino, T.; Takayama, S.; Koizumi, N. Characterization of a plant-specific gene induced by endoplasmic reticulum stress in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2010, 74, 2087–2091.
- Oslowski, C.M.; Urano, F. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2011; Volume 490, pp. 71–92.
- McCormack, M.E.; Liu, X.; Jordan, M.R.; Pajerowska-Mukhtar, K.M. An improved high-throughput screening assay for tunicamycin sensitivity in Arabidopsis seedlings. Front. Plant Sci. 2015, 6, 663.
- Koizumi, N.; Ujino, T.; Sano, H.; Chrispeels, M.J. Overexpression of a gene that encodes the first enzyme in the biosynthesis of asparagine-linked glycans makes plants resistant to tunicamycin and obviates the tunicamycin-induced unfolded protein response. Plant Physiol. 1999, 121, 353–362.
- Saijo, Y.; Tintor, N.; Lu, X.; Rauf, P.; Pajerowska-Mukhtar, K.; Häweker, H.; Dong, X.; Robatzek, S.; Schulze-Lefert, P. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 2009, 28, 3439–3449.
- Häweker, H.; Rips, S.; Koiwa, H.; Salomon, S.; Saijo, Y.; Chinchilla, D.; Robatzek, S.; von Schaewen, A. Pattern recognition receptors require N-glycosylation to mediate plant immunity. J. Biol. Chem. 2010, 285, 4629–4636.
- Deng, Y.; Srivastava, R.; Howell, S.H. Endoplasmic reticulum (ER) stress response and its physiological roles in plants. Int. J. Mol. Sci. 2013, 14, 8188–8212.
- Yu, X.; Wang, T.; Zhu, M.; Zhang, L.; Zhang, F.; Jing, E.; Ren, Y.; Wang, Z.; Xin, Z.; Lin, T. Transcriptome and physiological analyses for revealing genes involved in wheat response to endoplasmic reticulum stress. BMC Plant Biol. 2019, 19, 193.
- Rand, J.D.; Grant, C.M. The thioredoxin system protects ribosomes against stress-induced aggregation. Mol. Biol. Cell 2006, 17, 387–401.
- Altuntaş, C.; Terzi, R. Dithiothreitol and PEG Induced Combined Stress May Affect the Expressions of ABA Aldehyde Oxidase, Sucrose Synthase and Proline Metabolic Genes in Maize Seedlings. Phyton 2020, 89, 487.
- Howell, S.H. Endoplasmic reticulum stress responses in plants. Ann. Rev. Plant Biol. 2013, 64, 477–499.
- Li, B.; Yi, P.; Zhang, B.; Xu, C.J.; Liu, Q.Y.; Pi, Z.J.; Xu, X.L.; Chevet, E.; Liu, J.F. Differences in endoplasmic reticulum stress signalling kinetics determine cell survival outcome through activation of MKP-1. Cell Signal 2011, 23, 35–45.
- Srivastava, R.; Chen, Y.; Deng, Y.; Brandizzi, F.; Howell, S.H. Elements proximal to and within the transmembrane domain mediate the organelle-to-organelle movement of bZIP28 under ER stress conditions. Plant J. 2012, 70, 1033–1042.
- Li, Y.; Humbert, S.; Howell, S.H. ZmbZIP60 mRNA is spliced in maize in response to ER stress. BMC Res. Notes 2012, 5, 144.
- Moreno, A.A.; Mukhtar, M.S.; Blanco, F.; Boatwright, J.L.; Moreno, I.; Jordan, M.R.; Chen, Y.; Brandizzi, F.; Dong, X.; Orellana, A. IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS ONE 2012, 7, e31944.
- Bao, Y.; Bassham, D.C.; Howell, S.H. A functional unfolded protein response is required for normal vegetative development. Plant Physiol. 2019, 179, 1834–1843.
- Kim, J.-S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. ER-anchored transcription factors bZIP17 and bZIP28 regulate root elongation. Plant Physiol. 2018, 176, 2221–2230.
- Sun, L.; Yang, Z.T.; Song, Z.T.; Wang, M.J.; Sun, L.; Lu, S.J.; Liu, J.X. The plant-specific transcription factor gene NAC 103 is induced by b ZIP 60 through a new cis-regulatory element to modulate the unfolded protein response in A rabidopsis. Plant J. 2013, 76, 274–286.
- Liu, J.-X.; Howell, S.H. Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 2010, 22, 2930–2942.
- Urade, R. The endoplasmic reticulum stress signaling pathways in plants. Biofactors 2009, 35, 326–331.
- Neill, E.M.; Byrd, M.C.; Billman, T.; Brandizzi, F.; Stapleton, A.E. Plant growth regulators interact with elevated temperature to alter heat stress signaling via the Unfolded Protein Response in maize. Sci. Rep. 2019, 9, 10392.
- Carvalho, H.H.; Silva, P.A.; Mendes, G.C.; Brustolini, O.J.; Pimenta, M.R.; Gouveia, B.C.; Valente, M.A.S.; Ramos, H.J.; Soares-Ramos, J.R.; Fontes, E.P. The endoplasmic reticulum binding protein BiP displays dual function in modulating cell death events. Plant Physiol. 2014, 164, 654–670.
- Guan, P.; Wang, J.; Li, H.; Xie, C.; Zhang, S.; Wu, C.; Yang, G.; Yan, K.; Huang, J.; Zheng, C. Sensitive to SALT1, an endoplasmic reticulum-localized chaperone, positively regulates salt resistance. Plant Physiol. 2018, 178, 1390–1405.
- Henriquez-Valencia, C.; Moreno, A.A.; Sandoval-Ibañez, O.; Mitina, I.; Blanco-Herrera, F.; Cifuentes-Esquivel, N.; Orellana, A. bZIP17 and bZIP60 regulate the expression of BiP3 and other salt stress responsive genes in an UPR-independent manner in Arabidopsis thaliana. J. Cell. Biochem. 2015, 116, 1638–1645.
- Nawkar, G.M.; Kang, C.H.; Maibam, P.; Park, J.H.; Jung, Y.J.; Chae, H.B.; Chi, Y.H.; Jung, I.J.; Kim, W.Y.; Yun, D.-J. HY5, a positive regulator of light signaling, negatively controls the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 2084–2089.
- Kørner, C.J.; Du, X.; Vollmer, M.E.; Pajerowska-Mukhtar, K.M. Endoplasmic reticulum stress signaling in plant immunity—at the crossroad of life and death. Int. J. Mol. Sci. 2015, 16, 26582–26598.
- Liu, J.X.; Howell, S.H. Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol. 2016, 211, 418–428.
- Ron, D.; Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007, 8, 519.
- Angelos, E.; Ruberti, C.; Kim, S.J.; Brandizzi, F. Maintaining the factory: The roles of the unfolded protein response in cellular homeostasis in plants. Plant J. 2017, 90, 671–682.
- Angelos, E.; Brandizzi, F. NADPH oxidase activity is required for ER stress survival in plants. Plant J. 2018, 96, 1106–1120.
- Tateda, C.; Ozaki, R.; Onodera, Y.; Takahashi, Y.; Yamaguchi, K.; Berberich, T.; Koizumi, N.; Kusano, T. NtbZIP60, an endoplasmic reticulum-localized transcription factor, plays a role in the defense response against bacterial pathogens in Nicotiana tabacum. J. Plant Res. 2008, 121, 603–611.
- Pastor-Cantizano, N.; Ko, D.K.; Angelos, E.; Pu, Y.; Brandizzi, F. Functional Diversification of ER Stress Responses in Arabidopsis. Trends Biochem. Sci. 2020, 45, 123–136.
- Wang, L.; Mei, X.P.; Nan, J.; Liu, C.X.; Zhou, L.; Cai, Y.L. Overexpression of ZmNF-YC14 confers plant ER stress tolerance and ABA sensitivity in Arabidopsis. Acta Physiol. Plant. 2019, 41, 138.
- Liu, Y.; Burgos, J.S.; Deng, Y.; Srivastava, R.; Howell, S.H.; Bassham, D.C. Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 2012, 24, 4635–4651.
- Gupta, D.; Tuteja, N. Chaperones and foldases in endoplasmic reticulum stress signaling in plants. Plant Signal. Behav. 2011, 6, 232–236.
- Srivastava, R.; Li, Z.; Russo, G.; Tang, J.; Bi, R.; Muppirala, U.; Chudalayandi, S.; Severin, A.; He, M.; Vaitkevicius, S.I. Response to persistent ER stress in plants: A multiphasic process that transitions cells from Prosurvival activities to cell death. Plant Cell 2018, 30, 1220–1242.
- Moreno, A.A.; Orellana, A. The physiological role of the unfolded protein response in plants. Biol. Res. 2011, 44, 75–80.
- Alcântara, A.; Seitner, D.; Navarrete, F.; Djamei, A. A high-throughput screening method to identify proteins involved in unfolded protein response of the endoplasmic reticulum in plants. Plant Methods 2020, 16, 4.
- Ellgaard, L.; Helenius, A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003, 4, 181–191.
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science 2011, 334, 1081–1086.
- Gao, H.; Brandizzi, F.; Benning, C.; Larkin, R.M. A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2008, 105, 16398–16403.
- Srivastava, R.; Deng, Y.; Howell, S.H. Stress sensing in plants by an ER stress sensor/transducer, bZIP28. Front. Plant Sci. 2014, 5, 59.
- Römisch, K. Endoplasmic reticulum–associated degradation. Annu. Rev. Cell Dev. Biol. 2005, 21, 435–456.
- Park, J.H.; Kang, C.H.; Nawkar, G.M.; Lee, E.S.; Paeng, S.K.; Chae, H.B.; Chi, Y.H.; Kim, W.Y.; Yun, D.J.; Lee, S.Y. EMR, a cytosolic-abundant ring finger E3 ligase, mediates ER-associated protein degradation in Arabidopsis. N. Phytol. 2018, 220, 163–177.
- Hoseki, J.; Ushioda, R.; Nagata, K. Mechanism and components of endoplasmic reticulum-associated degradation. J. Biochem. 2010, 147, 19–25.
- Orenstein, S.J.; Cuervo, A.M. Chaperone-mediated autophagy: Molecular mechanisms and physiological relevance. Semin. Cell Dev. Biol. 2010, 21, 719–726.
- Yang, Z.; Klionsky, D.J. An overview of the molecular mechanism of autophagy. In Autophagy in Infection and Immunity; Springer: Berlin/Heidelberg, Germany, 2009; pp. 1–32.
- Mijaljica, D.; Prescott, M.; Devenish, R.J. Microautophagy in mammalian cells: Revisiting a 40-year-old conundrum. Autophagy 2011, 7, 673–682.
- Li, W.-w.; Li, J.; Bao, J.-k. Microautophagy: Lesser-known self-eating. Cell. Mol. Life Sci. 2012, 69, 1125–1136.
- Floyd, B.E.; Morriss, S.C.; MacIntosh, G.C.; Bassham, D.C. What to Eat: Evidence for Selective Autophagy in Plants F. J. Integr. Plant Biol. 2012, 54, 907–920.
- Bassham, D.C.; Laporte, M.; Marty, F.; Moriyasu, Y.; Ohsumi, Y.; Olsen, L.J.; Yoshimoto, K. Autophagy in development and stress responses of plants. Autophagy 2006, 2, 2–11.
- Hu, S.; Ye, H.; Cui, Y.; Jiang, L. AtSec62 is critical for plant development and is involved in ER-phagy in Arabidopsis thaliana. J. Integr. Plant Biol. 2020, 62, 181–200.
- Zhang, H.; Zhang, H. Autophagy: A self-eating mechanism for maintaining cellular homeostasis. Chin. Sci. Bull. 2016, 61, 3903–3906.
- Liu, Y.; Bassham, D.C. Autophagy: Pathways for self-eating in plant cells. Ann. Rev. Plant Biol. 2012, 63, 215–237.
- Pu, Y.; Bassham, D.C. Links between ER stress and autophagy in plants. Plant Signal. Behav. 2013, 8, e24297.




