
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Antonio J Marquez | + 3911 word(s) | 3911 | 2020-06-29 06:10:34 | | | |
| 2 | Catherine Yang | -64 word(s) | 3847 | 2020-07-31 05:54:11 | | |
Video Upload Options
Phenylpropanoid metabolism represents an important metabolic pathway from which originates a wide number of secondary metabolites derived from phenylalanine or tyrosine, such as flavonoids and isoflavonoids, crucial molecules in plants implicated in a large number of biological processes. Therefore, various types of interconnection exist between different aspects of nitrogen metabolism and the biosynthesis of these compounds. For legumes, flavonoids and isoflavonoids are postulated to play pivotal roles in adaptation to their biological environments, both as defensive compounds (phytoalexins) and as chemical signals in symbiotic nitrogen fixation with rhizobia. In this paper, we summarize the recent progress made in the characterization of flavonoid and isoflavonoid biosynthetic pathways in the model legume Lotus japonicus (Regel) Larsen under different abiotic stress situations, such as drought, the impairment of photorespiration and UV-B irradiation. Emphasis is placed on results obtained using photorespiratory mutants deficient in glutamine synthetase. The results provide different types of evidence showing that an enhancement of isoflavonoid compared to standard flavonol metabolism frequently occurs in Lotus under abiotic stress conditions. The advance produced in the analysis of isoflavonoid regulatory proteins by the use of co-expression networks, particularly MYB transcription factors, is also described. The results obtained in Lotus japonicus plants can be also extrapolated to other cultivated legume species, such as soybean, of extraordinary agronomic importance with a high impact in feeding, oil production and human health.
1. Introduction
The use of nitrogen by plants involves several steps, including uptake, assimilation, translocation, and different forms of recycling and remobilization processes, all of them of crucial importance in terms of nitrogen utilization efficiency. Different processes exist in plants, which give rise to the production of endogenous sources of ammonium which have to be efficiently re-assimilated by secondary ammonium assimilation. These processes include photorespiration, the biosynthesis of phenylpropanoids, as well as ureide, nucleotide and amino acid catabolism [1]. Phenylpropanoid metabolism represents an important metabolic pathway from which originates a wide number of secondary metabolites derived from phenylalanine or tyrosine, including monolignols, flavonoids and isoflavonoids, various phenolic acids, and stilbenes [2]. It is well known that secondary metabolites are crucial molecules in plant life, as protective agents against environmental factors (e.g., oxidative stress, pathogens, etc.) as well as elements favoring reproduction [3][4][5][6]. In particular, it is well established that phenylpropanoid-derived compounds have roles in plant growth and development, and in the defense against biotic and abiotic stress [2]. The phenylpropanoid pathway has different branches that lead to different families of compounds, such as chalcones, flavones, flavonols, flavanones, isoflavonoids, and anthocyanins, among others [7]. The structure, composition and biological activity of flavonoids have been frequently analyzed (see [7][8][9][10] for an overview, and references therein).
The second most important family of crop plants for humans, after Poaceae, are Fabaceae because they provide sources of food, feed for livestock and raw materials for industry [11]. Legumes are crucial plants in sustainable agriculture because they are able to fix atmospheric dinitrogen in a symbiotic association with rhizobial species. In addition, legumes produce a high diversity of secondary metabolites which serve as defense compounds against herbivores and microbes, but also as signal compounds to attract pollinating and fruit-dispersing animals. As nitrogen-fixing organisms, legumes produce more nitrogen containing secondary metabolites than other plant families [12]. In particular, flavonoids and isoflavonoids, which are compounds lacking nitrogen in their structures, are postulated to play pivotal roles in the adaptation of legumes to their biological environments both as defensive compounds (phytoalexins) and as chemical signals in symbiotic nitrogen fixation with rhizobia [13]. A primary function of flavonoids in legume–rhizobia symbiosis is to induce transcription of the genes involved in the biosynthesis of Nod factors. These factors are rhizobial signaling molecules perceived by the plant to allow symbiotic infection of the root. Many legumes produce specific flavonoids that only induce Nod factor production in compatible rhizobia, and therefore act as important determinants of host range [14]. Despite a wealth of evidence on legume flavonoids, relatively few have proven roles in rhizobial infection. The molecular details of how flavonoid production in plants is regulated during nodulation have not yet been clarified, but nitrogen availability has been shown to play a role [15]. The role of flavonoids and isoflavonoids in plant symbiosis is not limited to nitrogen-fixing bacteria since these compounds also play several roles in the symbiosis with mutualistic fungi. During the establishment of fungal symbiosis, these compounds can stimulate spore germination, hyphal branching and growth, root colonization, and arbuscule formation inside the root [16]. In later stages of symbiosis, flavonoids may be involved in the autoregulation of mycorrhization [17]. In the case of soybean, a specific isoflavonoid rather than a flavonoid can stimulate hyphal growth [18]. These effects often are host-specific, much like in the case of plant–rhizobial symbiosis. In fact, autoregulation of nodulation and autoregulation of mycorrhizae, the two negative feedback loops that control the formation of rhizobial and mycorrhizal symbioses, may share common elements [19]. However, the inhibitory effects of some plant flavonoids on fungal symbiosis have also been reported, both in plants that are host for mycorrhizal fungi and in non-host plants ([16], and references therein). Flavonoids can also accumulate in the early stages of plant–fungi interaction as a defense response; however, once the symbiosis has been established, the fungal symbiont may use the flavonoids as carbon source [20]. In addition, because legumes are a significant source of food and forage, the effects of leguminous flavonoids and isoflavonoids on human and animal health are being studied intensively [13][21]. In particular, excellent reviews describe exhaustively the different isoflavonoids compounds found in legume plants [22][23][24].
A major impetus in the investigation of the phenylpropanoid pathway in forage legumes was the fact that proanthocyanidins are beneficial in the diet of grazing ruminants through reduced pasture bloat, increase protein uptake and reduced intestinal parasite burdens [25][26]. Bloat is caused by protein foam formed in the rumen when animals graze protein-rich legume pastures. Rumen foam prevents normal expulsion of gases and, as consequence, ruminal volume and intraruminal pressure increase [26]. In the presence of proanthocyanidins, excess dietary proteins as well as bacterial enzymes are complexed and the level of protein degradation in the rumen is significantly reduced. This leads to an increased protein bypass to the ruminant’s gut and the improved absorption of essential amino acids, resulting in increased milk and meat production [25][26][27][28]. The production of pasture legume species with moderate amounts of foliar proanthocyanidins (2–4%) is of considerable interest to the pastoral agricultural industry [25][28][29].
Genomics and functional genomics, together with genetics, biochemistry, physiology, and molecular and cell biology, have accelerated discoveries in legume molecular and systems biology. Unfortunately, agricultural legumes are relatively poor model systems for research in genetics and genomics. Studies on most of the major leguminous crops are hampered by large genome sizes and other disadvantages (allogamy, polyploidy, transformation or regeneration recalcitrancies, few or large seeds and seedlings, genome duplications, long generation times, etc.). As a result, two species, Lotus japonicus and Medicago truncatula, were adopted internationally as models for modern legume research [30][31] and important advances have been produced in understanding the molecular details of rhizobial–legume symbiosis [32][33][34][35][36][37]. The high levels of synteny that exist between the different legume genomes imply that the advances obtained with the model plants can be used in order to understand and improve the performance of cultivated legume species [38].
In this paper, we will summarize recent progress made in the characterization of flavonoid and isoflavonoid biosynthetic pathways in legume plants with a particular focus on the model legume Lotus japonicus, and the impact that these studies may have to improve cultivated legumes of great agronomic importance such as soybean (Glycine max).
2. Flavonoid and Isoflavonoid Biosynthetic Pathways in Lotus
The enzyme chalcone synthase (CHS) is involved in the biosynthesis of the precursor molecules for both flavonoids and isoflavonoid biosynthesis. CHS is a member of the type III polyketide synthase family that catalyzes the conjugation of three acetate units from malonyl-CoA to a p-coumaroyl-CoA starter molecule derived from phenylalanine via the general phenylpropanoid pathway (Figure 1). In the same active site, additional aromatic “A” cycle of flavonoids is built via the intramolecular cyclisation [39]. The product of such reaction is 2’,4,4’,6’-tetrahydroxychalcone (naringenin chalcone), later changing to 5,7,4’-trihydroxyflavanone (naringenin) by building of the “C” heterocycle catalyzed by chalcone isomerase (CHI) that serves as a precursor for the other flavonoids [40]. In some species of the family Fabaceae, isoflavonoids, such as genistein, biochanin A or others, are produced from naringenin [41]. However, most of the isoflavonoids are synthesized via isoliquiritigenin that is produced by the coupled catalytic action of CHS and chalcone reductase (CHR; also called polyketide reductase, PKR, see below).
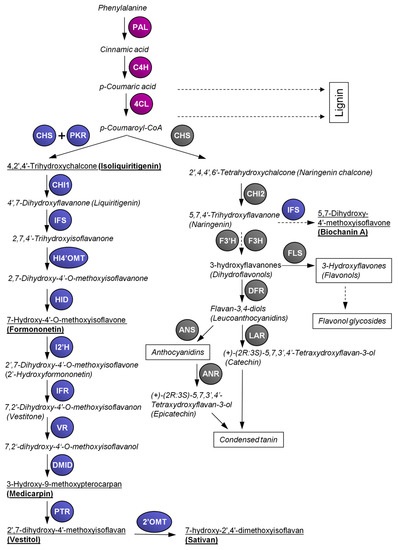
Figure 1. Overview of the flavonoid and isoflavonoid pathways in Lotus japonicus. 4CL, 4-coumarate:CoA ligase; 2’OMT, 2’-O-methyltransferase; I2’H, isoflavone-2’-hydroxylase; ANR, anthocyanidin reductase; ANS, anthocyanidin synthase; C4H, cinnamic acid 4-hydroxylase; CHI, chalcone isomerase; DMID, 7,2’-dihydroxy-4’-O-methoxyisoflavanol dehydratase (syn. pterocarpan synthase); CHS, chalcone synthase; DFR, dihydroflavonol 4-reductase; F3H, flavanone 3-hydroxylase; F3’H, flavanone 3’-hydroxylase; FLS, flavonol synthase; HID, 2-hydroxyisoflavanone dehydratase; HI4’OMT, 2-hydroxyisoflavanone 4’-O-methyltransferase; IFR, isoflavone reductase; IFS, 2-hydroxyisoflavanone synthase; LAR, leucoanthocyanidin reductase; PAL, phenylalanine ammonia lyase; PKR, polyketide reductase (syn. chalcone reductase); PTR, pterocarpan reductase; VR, vestitone reductase. Purple color: enzymes of general phenylpropanoid pathway; grey color: enzymes of flavonoid pathway; blue color: enzymes of isoflavonoid pathway. Dashed arrows represent multiple biosynthetic steps. Trivial names of compounds are presented if they are commonly used; the others are presented by their semi-systematic names. Semi-systematic names and chemical structures of the referred flavonoids and isoflavonoids . The names underlined in bold highlight most abundant isoflavonoids found in L. japonicus according to our data [42].
Whereas in Arabidopsis thaliana only one single gene for CHS is known, in other species several CHS genes were found (e.g., two in cacao, four in wild strawberry, five in apple, six in poplar), which is especially true for legumes [43][44]. In L. japonicus 13–14 CHS genes were found, 15 in Glycine max and 17 in Medicago truncatula [43]. The higher number of CHS genes in legumes is likely related to the presence of the isoflavonoid pathway in that family. In L. japonicus, CHS6 (called LjCHS1 in [45]) could represent the non-leguminous type of chalcone synthase; on the other hand, in soybean, GmCHS6, GmCHS7 and GmCHS8 seem more related to isoflavonoid production [46][47]. GmCHS7 and GmCHS8 show strong homology with LjCHS5 (Lj1g3v2626200.1), LjCHS8 (Lj0g3v0129339.1) LjCHS9 (Lj2g3v2124320.1) and LjCHS11 (Lj2g3v2124320.2), whereas GmCHS6 is homologous to LjCHS12 (Lj4g3v2574990.1). However, Lotus isoflavonoids are produced mainly via isoliquiritigenin, the daidzein and genistein (and their derivates) found in soybean are produced from isoliquiritigenin and naringenin chalcone, respectively [41][48] (Figure 1). Therefore, the regulation pattern of chalcone synthases in soybean might be more complex.
The flavonoid biosynthetic pathway producing flavonols, anthocyanidins and proanthocyanidins (condensed tannins) in L. japonicus are described in Figure 1. F3H, F3’H and FLS genes have not been studied in detail to date—five DFR genes were described in a cluster on chromosome 5 by [49]and different specificities of DFR isozymes in the substrate hydroxylation patterns have been reported. The proanthocyanidins (both epicatechin and catechin type) are biosynthesized from dihydroflavonols by the action of anthocyanidin reductase (ANR) and leucoanthocyanidin 4-reductase (LAR), two gene encodings for enzymes committed to epicatechin and catechin biosynthesis, respectively, that were identified in L. corniculatus [50].
Higher plants share a common core flavonoid pathway, although different species frequently develop specific branches as an adaptation to diverse environmental conditions. For example, A. thaliana accumulates mainly flavonols (kaempferol, quercetin and isorhamnetin glycosides) in all tissues, and anthocyanidins and epicatechin types of proanthocyanidins in the seed coat under stress conditions [51]. A rising number of studies report protein–protein interactions of flavonoid biosynthetic enzymes providing evidence for weakly bound complexes called “metabolons” which are co-localized at the endoplasmic reticulum (ER) [52][53][54]. The interaction of the enzymes in the system likely allows better connection of reaction intermediates with subsequent enzymes and prevents their loss by diffusion or unfavorable cell equilibrium. Such protein–protein interactions were found for CHS, flavanone 3-hydroxylase (F3H), dihydroxyflavonol 4-reductase (DFR), anthocyanidin synthase (ANS) and also CHI or CHI-like protein (with a putative role as fatty-acid binding protein) [55], so the proposed model of metabolon comprises the enzymes necessary for formation of anthocyanidins [56]. On the other hand, there is still lack of evidence of interaction with flavanone 3’–hydroxylase (F3’H) [57]. Proanthocyanidins are produced by action of ANR, LAR and polyphenol oxidase (LAC15) resulting in the oligo-and polymers of the flavan-3-ol units. Substrate channeling between DFR and LAR was described using molecular modeling and predicted the functional significance of metabolon formation during synthesis [58]. Proanthocyanidins are produced both in shoots and roots of Lotus sp. However, significant differences in their accumulation may occur among different species, but also within different populations of the same species. Whereas in L. japonicus (and some other species) they are usually present in almost undetectable amounts, the closely related tetraploid forage species L. corniculatus may accumulate proanthocyanidins in considerable levels [59][60]. The highest proanthocyanidin levels were found in L. unifoliolatus (syn. L. americanus) and L. uliginosus (syn. L. pedunculatus) [[59][61].
The key enzyme for flavonol formation is flavonol synthase (FLS) using dihydroflavonol substrates. L. japonicus is a plant that accumulates flavonol kaempferol glycosides in considerable amounts, especially kaempferol-3,7-dirhamnoside. Quercetin glycosides are present at lower levels but increase under some abiotic stress conditions [62][63]. Moreover, a considerable amount of gossypetine glycosides occurs in flowers and a small amount of isorhamnetine can be detected in stems [64]. Only the minor methylation on 3’ position of quercetine is present in L. japonicus, whereas the methylation at position 8 was described only in L. corniculatus [65], leading to presence of sexangularetin and corniculatusin in that species [66].
The production of isoliquiritigenin, the starting point of the second branch of the biosynthetic pathway, is related to the activity of CHR (also called polyketide reductase, PKR), only identified in papilionoid legumes (like Glycine max, Medicago sativa, Glycyrrhiza echinata, Glycyrrhiza glabra). Five genes and 1 pseudogene are present in the L. japonicus genome [35]. CHR acts in a coupled catalytic action with CHS [45]. Furthermore, two types of CHI genes are present. LjCHI2 is highly homologous to non-legumes (also referred as type I), whereas LjCHI1, LjCHI3 and LjCHI4 are legume-specific type II, also occurring in Medicago sativa, Phaseolus vulgaris, Pisum sativum and Pueraria lobata [67] (Figure 1). The legume-specific type II evolved to produce 5-deoxy(iso)flavonoids from 6’deoxychalcone (isoliquiritigenin) along with the establishment of the Fabaceae.
The protein–protein interaction of key enzymes of isoflavonoid pathway (CHS, CHR, CHI and IFS) that are associated with ER via cytochrome P450 has been recently demonstrated in soybean [68] as well as with the three enzymes of general phenylpropanoid pathway (PAL, C4H, 4CL) and with the last enzyme of the shikimate pathway, arogenate dehydratase (ADT), the enzyme converting arogenate to phenylalanine [69]. The enzyme complex may be associated with the ER membrane at the plastid-associated membrane sites, allowing the flux of intermediates from shikimate pathway occurring in plastids toward daidzein or glycitein isoflavones present in soybean [69][70].
Isoflavone synthase (IFS; 2-hydroxyisoflavanone synthase) is a membrane-associated enzyme belonging to the CYP93C subfamily of cytochrome P450 monooxygenases that constructs the isoflavonoid skeleton from 4’,7-dihydroxyflavanone substrate (liquiritigenin) by an unusual aryl migration reaction. At a lower rate, IFS may convert naringenin in several legume species, such as soybean [41]. IFS has been identified almost exclusively in legumes, with Beta vulgaris being the only known exception [71][72]. Among 273 putative P450 genes in A. thaliana genome, none of them has isoflavone synthesizing activity [73]. At least two functional genes of IFS (IFS1 and IFS2) and one pseudogene are present in the L. japonicus genome [45]. L. japonicus IFS likely has a strong preference for liquiritigenin, although a small amount of biochanin A detected in plants on UV-B irradiation suggests a possibility of a minor activity using also naringenin as a substrate [42].
The substrate specificity of 2-hydroxyisoflavanone dehydratase (HID) may differ among species. In soybean, HID accepts 2,5,7,4’-tetrahydroxyisoflavanone or 2,7,4’-trihydroxyisoflavanone as substrate, which is then de-hydrated to produce a double bond between C-2 and C-3, yielding genistein or daidzein [23][74]. The overexpression of HID from soybean with broad substrate specificity in L. japonicus resulted in the production of considerable amounts of daidzein or genistein [75]. The biosynthesis of the main isoflavonoid, vestitol, in L. japonicus was proposed by [45], according the previous data described in Glycyrrhiza echinata [74]. Firstly the 4’-O-methyltransferase (HI4‘OMT) reaction occurs, and subsequent dehydration by HID yields formononetin (Figure 1), the central biosynthetic intermediate for the production of diverse isoflavonoid phytoalexins (e.g., maackiain, pisatin, medicarpin, etc.) in a number of legume species, including agronomically important ones such as pea (Pisum sativum) or chickpea (Cicer arietinum) [76].
In L. japonicus, formononetin is then converted by isoflavone-2’-hydroxylase (I2’H) to 2’,7-dihydroxy-4’-O-methoxyisoflavone and subsequently to vestitone by isoflavone reductase (IFR). The next step is the NADPH-dependent reduction of vestitone to 7,2’-dihydroxy-4’-O-methoxyisoflavanol, catalyzed by the vestitone reductase (VR) that is stereospecific for the (3S)-vestitone [77]. HI4’OMT, HID and I2’H were suggested to occur in single copies in the L. japonicus genome [78], but recently, more putative copies could be predicted at least in the case of HID (miyakoguza.jp 3.0). The putative L. japonicus IFR1 and VR1, VR2, VR3 and VR4 genes (four VR genes) for vestitol accumulation were identified by sequence similarity with Medicago sativa [45]. Although their functional validation is still lacking, these genes are markedly upregulated after glutathione treatment [79]. The production of medicarpin from 7,2’-dihydroxy-4’-O-methoxyisoflavanol is catalyzed by pterocarpan synthase (PTS) that was found in L. japonicus, Glycine max and Glycyrrhiza echinata. This enzyme has similar biochemical properties as previously reported DMI-dehydratase in Cicer arientinum, G. max and Medicago sativa. This raises the question of whether the product of the LjPTS1 gene corresponds to the enzyme described above, but the evidence available at present is not conclusive [80]. The synthesis of vestitol is then catalyzed by pterocarpan reductase (PTR). Four genes were found to encode PTR, from which PTR3 was found to be inducible by glutathione [45]. However, PTR1 and PTR2 have much higher activity and enantiospecifity with (-)-medicarpin; therefore, they are considered to be responsible for vestitol production [81]. In stress conditions like UV-B application or glutathione treatment, a remarkable accumulation of sativan was observed in L. japonicus and L. corniculatus [42][82]. Production of sativan requires the activity of a 2’-O-methyltransferase to convert vestitol to sativan. Among the type I O-methyltransferases isolated from Medicago truncatula, MtOMT2, MtOMT4, MtOMT5, MtOMT6 and MtOMT7 showed some vestitol methylation activity, but with a very low efficiency. Furthermore, they appeared to methylate vestitol at the positions 7 and/or 4’; any clear evidence of methylation at 2’ position of vestitol is still lacking [83]. Vestitol is a predominant isoflavonoid produced in L. japonicus, present in very small amount in unstressed conditions, but increases significantly at biotic [84][85] or abiotic stresses [62] or after treatment with 10 mM glutathione [78][86]. To a lesser extent, sativan also accumulates in such conditions. Other isoflavonoids, such as formononetin and biochanin A, were raised after UV-B irradiation but their levels remained more than ten-times lower in comparison to vestitol. Accumulation of sativan and medicarpin was also detected, but in an even lower extent [42].
Glycosylation is a major decorative modification that occurs frequently as a last step of the biosynthesis of certain flavonoids or isoflavonoids. UDP sugar residues are attached to the flavonoid core via a uridine diphosphate glycosyltransferase (UGT) [87]. A large number of putative UGT genes have been identified in several plant species. However, only few of them were functionally characterized, mostly in Arabidopsis thaliana [88]. In the L. japonicus genome, 188 putative UGT genes were identified by genome-wide searching [89]. Tree UGT proteins of the UGT72 family enzymes (UGT72AD1, UGT72AH1 and UGT72Z2) showed narrow substrate preferences to flavonol aglycones in vitro and the overexpression of UGT72AD1 and UGT72Z2 led to increase of flavonol rhamnosides. Another two proteins, UGT72AF1 and UGT72V3, exhibited a broad activity towards flavonoids and isoflavonoids [89]. Such a broad activity of UGTs is known also from other legumes, in particular in the case of four UGTs (GT22D, GT22E09, GT29C and GT29H) from M. truncatula [90] and three UGTs (UGT73F2, UGT73C20 and UGT88E19) from G. max [91][92]. The UGT activity resulted to high diversity of glycosides in L. japonicus; particularly (25) kaempferol and (12) quercetine glycosides were found mostly in flowers [64].
3. Conclusions and Future Prospects
It summarizes recent advances made in flavonoid and isoflavonoid research in the model legume L. japonicus. The study of the response of L. japonicus to abiotic stress conditions led to different novel findings, such as the accumulation of new flavonols that were described for the first time in L. japonicus leaves [62] and of a peculiar pattern of isoflavonoid accumulation in the response of this plant to UV-B irradiation [42]. Despite the fact that flavonoid and isoflavonoid metabolism is a very active field of research; several aspects of these pathways are far from having been completely described. Technical advances in metabolomics are enabling the discovery of a growing number of flavonoids and isoflavonoids structures. However, chemical modification of the flavonoid/isoflavonoid scaffolds, such as glycosylation and acylation, add another layer of complexity to their chemical diversity; and the reason beyond such complexity is still not completely understood. Legumes also use flavonoids or isoflavonoids in order to attract their chosen symbiont in a species-specific way. Despite the important role played by L. japonicus in elucidating the molecular genetics of legume–rhizobia symbiosis, it is still unknown which class of phenolic compounds are used by this species in order to attract its chosen symbiont [15]. Studies of the symbiotic capacity of specific L. japonicus mutants impaired in specific branches of the biosynthesis of phenolic compounds, paired with metabolite profiling will be needed in order to fill this gap. The regulation of isoflavonoid metabolism is also far from being completely understood. A few negative and positive regulators have been identified in soybean, while no clear isoflavonoid regulators have been identified in L. japonicus to date. The co-expression analysis presented in this paper identified potential candidates for isoflavonoid regulation in L. japonicus. Future works should be aimed to the characterization of specific mutants in these genes in order to understand whether they are involved in isoflavonoid regulation, and also if they may play a role in the response to different kinds of abiotic stress. A deeper understanding of isoflavonoid regulation may also permit tackling the genetic improvement of soybean and to breed varieties with either increased or decreased isoflavonoid content, two opposite traits that can be desirable depending on the products that will be manufactured using these soya beans. Since most of the regulators identified in these species are from the MYB transcription factor family, which is composed of a very high number of genes, traditional approaches, such as searching for isoflavonoid-related QTL, may be very time consuming. Bioinformatics approaches, such as the construction and analysis of gene co-expression networks in order to find new candidate regulators, combined with validation of these genes by characterizing loss-of–function mutants, have already showed promising results. Finally, as explained in this review, in order to broaden the knowledge of flavonoid and isoflavonoid metabolism and regulation, studies that take into consideration both model species such as L. japonicus, of easier genetic manipulation, and cultivated species of great economic importance, such as soybean, will be of paramount impact for legume flavonoid/isoflavonoid research.
References
- Liu, J.; Jiang, W.; Xia, Y.; Wang, X.; Shen, G.; Pang, Y. Genistein-specific G6DT gene for the inducible production of wighteone in Lotus japonicus. Plant Cell Physiol. 2018, 59, 128–141.
- Betti, M.; García-Calderón, M.; Pérez-Delgado, C.M.; Credali, A.; Pal’ove-Balang, P.; Estivill, G.; Repçák, M.; Vega, J.M.; Galván, F.; Márquez, A.J. Reassimilation of ammonium in Lotus japonicus. J. Exp. Bot. 2014, 19, 5557–5566.
- Liu, J.; Osbourn, A.; Ma, P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 2015, 8, 689–708.
- Theis, N.; Lerdau, M. The evolution of function in plant secondary metabolites. Int. J. Plant Sci. 2003, 164, S93–S102.
- Izhaki, I. Emodin—A secondary metabolite with multiple ecological functions in higher plants. New Phytol. 2002, 155, 205–217.
- De Geyter, N.; Gholami, A.; Goormachtig, S.; Goossens, A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 2012, 17, 349–359.
- Novelli, S.; Gismondi, A.; Di Marco, G.; Canuti, L.; Nanni, V.; Canini, A. Plant defense factors involved in Olea europea resistance against Xylella fastidiosa infection. J. Plant Res. 2019, 132, 439–455.
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47.
- Falcone Ferreyra, M.L.; Rius, S.P.; Casati, D.P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222.
- Tohge, T.; Perez de Souza, L.; Fernie, A.R. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 2017, 68, 4013–4028.
- Mathesius, U. Flavonoid functions in plants and their interactions with other organisms. Plants 2018, 7, 30.
- Graham, P.H.; Vance, C.P. Legumes: Importance and constraints to greater use. Plant Physiol. 2003, 131, 872–877.
- Wink, M. Evolution of secondary metabolites in legumes (Fabaceae). S. Afr. J. Bot. 2013, 89, 164–175.
- Aoki, T.; Akashi, T.; Ayabe, S. Flavonoids of leguminous plants: Structure, biological activity, and biosynthesis. J. Plant Res. 2000, 113, 475–488.
- Weston, L.A.; Mathesius, U. Flavonoids: Their structure, biosynthesis and role in the rhizosphere, including allelopathy. J. Chem. Ecol. 2013, 39, 283–297.
- Liu, C.; Murray, J.D. The role of Flavonoids in Nodulation Host-Range Specificity: An update. Plants 2016, 5, 33.
- Hassan, S.; Mathesius, U. The role of flavonoids in root-rhizosphere signaling: Opportunities and challenges for improving plant-microbe interactions. J. Exp. Bot. 2012, 63, 3429–3444.
- Larose, G.; Chenevert, R.; Moutuglis, P.; Gagne, S.; Piche, Y.; Vierheilig, H. Flavonoid levels in Medicago sativa are modulated by the developmental stage of the symbiosis and the root colonizing arbuscular mycorrhizal fungus. J. Plant Physiol. 2002, 159, 1329–1339.
- Morandi, D.; Bailey, J.A.; Gianinazzi-Pearson, V. Isoflavonoid accumulation in soybean roots infected with vesicular-arbuscular mycorrhizal fungi. Physiol. Plant Pathol. 1984, 24, 357–364.
- Wang, C.; Reid, J.B.; Foo, E. The art of self-control—Autoregulation of plant-microbe symbioses. Front. Plant Sci. 2018, 9, 988.
- Harrison, M.; Dixon, R.A. Isoflavonoid accumulation and expression of defense gene transcripts during the establishment of vescicular-arbuscular mycorrhizal associations in roots of Medicago truncatula. Mol. Plant Microbe Interact. 1993, 6, 643–654.
- Dwivedi, S.L.; Upadhyaya, H.D.; Chung, I.-M.; De Vita, P.; García-Lara, S.; Guajardo-Flores, D.; Gutiérrez-Uribe, J.A.; Serna-Saldívar, S.O.; Rajakumar, G.; Sahrawat, K.L.; et al. Exploiting phenylpropanoid derivatives to enhance the nutraceutical values of cereals and legumes. Front. Plant Sci. 2016, 7, 763.
- Veitch, N.C. Isoflavonoids of the leguminosae. Nat. Prod. Rep. 2007, 24, 417–464.
- Veitch, N.C. Isoflavonoids of the Leguminosae. Nat. Prod. Rep. 2009, 26, 776–802.
- Veitch, N.C. Isoflavonoids of the leguminosae. Nat. Prod. Rep. 2013, 30, 988–1027.
- Aerts, R.J.; Barry, T.N.; McNabb, W.C. Polyphenols and agriculture: Beneficial effects of proanthocyanidins in forages. Agric. Ecosyst. Environ. 1999, 75, 1–12.
- McMahon, L.R.; McAllister, T.A.; Berg, B.P.; Majak, W.; Acharya, S.N.; Popp, J.D.; Coulman, B.E.; Wang, Y.; Cheng, K.-J. A review of the effects of forage condensed tannins on ruminal fermentation and bloat in grazing cattle. Can. J. Plant Sci. 2000, 80, 469–485.
- Douglas, G.B.; Stienezen, M.; Waghorn, G.C.; Foote, A.G.; Purchas, R.W. Effect of condensed tannins in birdsfoot trefoil (Lotus corniculatus) and sulla (Hedysarum coronarium) on body weight, carcass fat depth, and wool growth of lambs in New Zealand. N. Z. J. Agric. Res. 1999, 42, 55–64.
- Hancock, KR.; Collette, V.; Fraser, K.; Greig, M.; Xue, H.; Richardson, K.; Jones, C.; Rasmussen, S. Expression of the R2R3-MYB Transcription factor TaMYB14 from Trifolium arvense activates proanthocyanidin biosynthesis in the legumes Trifolium repens and Medicago sativa. Plant Physiol. 2012, 159, 1204–1220.
- Jonker, A.; Yu, P. The role of proanthocyanidins complex in structure and nutrition interaction in alfalfa forage. Int. J. Mol. Sci. 2016, 17, 793.
- Márquez, A.J. (Ed.) Lotus japonicus Handbook, 1st ed.; Springer: Dordrecht, The Netherlands, 2005; pp. 1–384.
- Mathesius, U.; Journet, E.P.; Sumner, L.W. (Eds.) The Medicago truncatula Handbook. Available online: http://www.noble.org/medicago-handbook/ (accessed on 28 November 2019).
- Stacey, G.; Libault, M.; Brechenmacher, L.; Wan, J.; May, G.D. Genetics and functional genomics of legume nodulation. Curr. Opin. Plant Biol. 2006, 9, 110–121.
- Udvardi, M.K.; Tabata, S.; Parniske, M.; Stougaard, J. Lotus japonicus: Legume research in the fast lane. Trends Plant Sci. 2005, 10, 222–228.
- Madsen, L.H.; Tirichine, L.; Jurkiewicz, A.; Sullivan, J.T.; Heckman, A.B.; Bek, A.S.; Ronson, C.W.; James, E.K.; Stougaard, J. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 2010, 1, 10.
- Oldroyd, G.E.D.; Murray, J.D.; Poole, P.S.; Downie, J.A. The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 2011, 45, 119–144.
- Oldroyd, G.E.D. Speak, friend, and enter: Signalling systems that promote symbiotic associations in plants. Nat. Rev. Microbiol. 2013, 11, 252–263.
- Udvardi, M.; Poole, P.S. Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 2013, 64, 781–805.
- Libault, M.; Joshi, T.; Benedito, V.A.; Xu, D.; Udvardi, M.K.; Stacey, G. Legume transcription factor genes: What makes legumes so special? Plant Physiol. 2009, 151, 991–1001.
- Austin, M.B.; Noel, J.P. The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 2003, 20, 79–110.
- Winkel-Shirley, B. It takes a garden. How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiol. 2001, 127, 1399–1404.
- Yerlikaya, S.; Baloglu, M.C.; Diuzheva, A.; Jekő, J.; Cziáky, Z.; Zengin, G. Investigation of chemical profile, biological properties of Lotus corniculatus L. extracts and their apoptotic-autophagic effects onbreast cancer cells. J. Pharm. Biomed. Anal. 2019, 174, 286–299.
- Cheng, H.; Wang, J.; Chu, S.; Yan, H.-L.; Yu, D. Diversifying selection on flavanone 3-hydroxylase and isoflavone synthase genes in cultivated soybean and its wild progenitors. PLoS ONE 2013, 8, e54154.
- Zavala, K.; Opazo, J.C. Lineage-specific expansion of the chalcone synthase gene family in rosids. PLoS ONE 2015, 10, e0133400.
- Dao, T.T.H.; Linthorst, H.J.M.; Verpoorte, R. Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 2011, 10, 397–412.
- Shimada, N.; Sato, S.; Akashi, T.; Nakamura, Y.; Tabata, S.; Ayabe, S.-I.; Aoki, T. Genome-wide analyses of the structural gene families involved in the legume-specific 5-deoxyisoflavonoid biosynthesis of Lotus japonicus. DNA Res. 2007, 14, 25–36.
- Daubhadel, S.; Gijyen, M.; Moy, P.; Farhangkhoee, M. Transcriptome analysis reveals a critical role of CHS7 and CHS8 genes for isoflavonoid synthesis in soybean seeds. Plant Physiol. 2007, 143, 326–338.
- Lim, J.Y.; Jeon, H.Y.; Gil, Ch.S.; Kwon, S.-J.; Na, J.K.; Lee, Ch.; Eom, S.H. Isoflavone accumulation and the metabolic gene expression in response to persistent UV-B irradiation in soybean sprouts. Food Chem. 2020, 303, 125376.
- Masunaka, A.; Hyakumachi, M.; Takenaka, S. Plant growth–promoting fungus, Trichoderma koningi suppresses isoflavonoid phytoalexin vestitol production for colonization on/in the roots of Lotus japonicus. Microbes Environ. 2011, 26, 128–134.
- Shimada, N.; Sasaki, R.; Sato, S.; Kneko, T.; Tabata, S.; Aoki, T.; Ayabe, S.-I. A comprehensive analysis of six dihydroflavonol 4-reductases encoded by a gene cluster of the Lotus japonicus genome. J. Exp. Bot. 2005, 56, 2573–2585.
- Paolocci, F.; Robbins, M.P.; Madeo, L.; Arcioni, S.; Martens, S.; Damiani, F. Ectopic expression of a basic helix-loop-helix gene transactivates parallel pathways of proanthocyanidin biosynthesis. Structure, expression analysis, and genetic control of leucoanthocyanidin 4-reductase and anthocyanidin reductase genes in Lotus corniculatus. Plant Physiol. 2007, 143, 504–516.
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 2013, 72, 21–34.
- Saslowsky, D.; Winkel-Shirley, B. Localization of flavonoid enzymes in Arabidopsis roots. Plant J. 2001, 27, 37–48.
- Crosby, K.C.; Pietraszewska-Bogiel, A.; Gadella, T.W.J.; Winkel, B.S.J. Förster resonance energy transfer demonstrates a flavonoid metabolon in living plant cells that displays competitive interactions between enzymes. FEBS Lett. 2011, 585, 2193–2198.
- Sweetlove, L.J.; Fernie, A.R. The spatial organization of metabolism within the plant cell. Annu. Rev. Plant Biol. 2013, 64, 723–746.
- Ngaki, M.N.; Louie, G.V.; Philippe, G.; Manning, G.; Pojer, F.; Bowman, M.E.; Li, L.; Larsen, E.; Wurtele, E.S.; Noel, J.P. Evolution of the chalcone-isomerase fold from fatty-acid binding to stereospecific catalysis. Nature 2012, 485, 530–533.
- Nakayama, T.; Takahashi, S.; Waki, T. Formation of flavonoid metabolons: Functional significance of protein-protein interactions and impact on flavonoid chemodiversity. Front. Plant Sci. 2019, 10, 821.
- Ralston, L.; Yu, O. Metabolons involving plant cytochrome P450s. Phytochem. Rev. 2006, 5, 459–472.
- Diharce, J.; Golebiowski, J.; Fiorucci, S.; Antonczak, S. Fine-tuning of microsolvation and hydrogen bond interaction regulates substrate channelling in the course of flavonoid biosynthesis. Phys. Chem. Chem. Phys. 2016, 18, 10337–10345.
- Gruber, M.; Skadhauge, B.; Yu, M.; Muir, A.; Richards, K. Variation in morphology, plant habit, proanthocyanidins, and flavonoids within a Lotus germplasm collection. Can. J. Plant Sci. 2008, 88, 121–132.
- Escaray, F.J.; Passeri, V.; Babuin, F.M.; Marco, F.; Carrasco, P.; Damiani, F.; Pieckenstain, F.L.; Poalocci, F.; Ruiz, O. Lotus tenuis x L. corniculatus interspecific hybridization as a means to breed bloat-safe pastures and gain insight into the genetic control of proanthocyanidin biosynthesis in legumes. BMC Plant Biol. 2014, 14, 40.
- Sivakumaran, S.; Rumball, W.; Lane, G.F.; Fraser, K.; Foo, L.Y.; Yu, M.; Meagher, L.P. Variation of proanthocyanidins in Lotus species. J. Chem. Ecol. 2006, 32, 1797–1816.
- García-Calderón, M.; Pons-Ferrer, T.; Mrázová, A.; Pal’ove-Balang, P.; Vilková, M.; Pérez-Delgado, C.M.; Vega, J.M.; Eliášová, A.; Repčák, M.; Márquez, A.J.; et al. Modulation of phenolic metabolism under stress conditions in a Lotus japonicus mutant lacking plastidic glutamine synthetase. Front. Plant Sci. 2015, 6, 760.
- Mrázová, A.; Mrázová, A.; Belay, S.A.; Eliášová, A.; Perez-Delgado, C.; Kaducová, M.; Betti, M.; Vega, J.M.; Pal’ove-Balang, P. Expression, activity of phenylalanine-ammonia-lyase and accumulation of phenolic compounds in Lotus japonicus under salt stress. Biologia 2017, 72, 36–42.
- Suzuki, H.; Sasaki, R.; Ogata, Y.; Nakamura, Y.; Sakurai, N.; Kitajima, M.; Takayama, H.; Kanaya, S.; Aoki, K.; Shibata, D.; et al. Metabolic profiling of flavonoids in Lotus japonicus using liquid chromatography Fourier transform ion cyclotron resonance mass spectrometry. Phytochemistry 2008, 69, 99–111.
- Jay, M.; De Luca, V.; Ibrahim, R. Meta-methylation of flavonol rings A (8-) and B (3’-) is catalysed by two distinct O-methyltransferases in Lotus corniculatus. Z. Nat. C 1983, 38, 413–417.
- Kaducová, M.; Monje-Rueda, M.D.; García-Calderón, M.; Pérez-Delgado, M.C.; Eliášová, A.; Gajdošová, S.; Petruľová, V.; Betti, M.; Márquez, A.J.; Paľove-Balang, P. Induction of isoflavonoid biosynthesis in Lotus japonicus after UV-B irradiation. J. Plant Physiol. 2019, 236, 88–95.
- Shimada, N.; Aoki, T.; Sato, S.; Nakamura, Y.; Tabata, S.; Ayabe, S.-I. A cluster of genes encodes the two types of chalcone isomerase involved in the biosynthesis of general flavonoids and legume-specific 5-deoxy(iso)flavonoids in Lotus japonicus. Plant Physiol. 2003, 131, 941–951.
- Waki, T.; Yoo, D.-C.; Fujino, N.; Mameda, R.; Dennesiouk, K.; Yamashita, S.; Motohashi, R.; Akashi, T.; Aoki, T.; Ayabe, S.-I.; et al. Identification of protein protein interactions of isoflavonoid biosynthetic enzymes with 2-hydroxyisoflavanone synthase in soybean (Glycine max (L.) Merr.). Biochem. Biophys. Res. Commun. 2016, 469, 546–551.
- Dastmalchi, M.; Bernards, M.A.; Dhaubhadel, S. Twin anchors of the soybean isoflavonoid metabolon: Evidence for tethering of the complex to the endoplasmic reticulum by IFS and C4H. Plant J. 2016, 85, 689–706.
- Winkel, B. The Subtleties of Subcellular Distribution. Pointing the way to underexplored functions for flavonoid enzymes and end products. In Recent Advances in Polyphenol Research; Halbwirth, H., Stich, K., Cheynier, V., Quideau, S., Eds.; Jonh Willey & Sons. Ltd.: Chichester, UK, 2019; Volume 6, pp. 89–108.
- Jung, W.; Yu, O.; Cindy Lau, S.-M.; O’Keefe, D.P.; Odell, J.; Fader, G. McGonigle, B. Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat. Biotechnol. 2000, 18, 208–212.
- Lapčík, O. Isoflavonoids in non-leguminous taxa: A rarity or a rule? Phytochemistry 2007, 68, 2909–2916.
- Wang, X. Structure, function and engineering of enzymes in isoflavonoid biosynthesis. Funct. Integr. Genom. 2011, 11, 13–22.
- Akashi, T.; Aoki, T.; Ayabe, S.-I. Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of carboxylesterase-like proteins in leguminous isoflavone biosynthesis. Plant Physiol. 2005, 137, 882–891.
- Shimamura, M.; Akashi, T.; Sakurai, N.; Suzuki, H.; Saito, K.; Shibata, D.; Ayabe, S.-I.; Aoki, T. 2-Hydroxyisoflavanone dehydratase is a critical determinant of isoflavone productivity in hairy root cultures of Lotus japonicus. Plant Cell Physiol. 2007, 48, 1652–1657.
- Jeandet, P.; Clément, E.; Courot, E.; Cordelier, S. Modulation of phytoalexin biosynthesis in engineered plants for disease resistance. Int. J. Mol. Sci. 2013, 14, 14136–14170.
- Shao, H.; Dixon, R.A.; Wang, X. Crystal structure of vestitone reductase from alfalfa (Medicago sativa L.). J. Mol. Biol. 2007, 369, 265–276.
- Shimada, N.; Akashi, T.; Aoki, T.; Ayabe, S.i. Induction of isoflavonoids pathway in the model legume Lotus japonicus: Molecular characterization of enzymes involved in phytoalexin biosynthesis. Plant Sci. 2000, 160, 37–47.
- Monje-Rueda, M.D.; García-Calderón, M.; Pal’Ove-Balang, P.; Márquez, A.J.; Betti, M. Functional analysis of two Lotus japonicus mutants affected in MYB transcription factors. (manuscript in preparation).
- Uchida, K.; Akashi, T.; Aoki, T. The missing link in leguminous pterocarpan biosynthesis is a dirigent domain-containing protein with isoflavanol dehydratase activity. Plant Cell Physiol. 2017, 58, 398–408.
- Akashi, T.; Koshimizu, S.; Aoki, T.; Ayabe, S.-I. Identification of cDNA encoding pterocarpan reductase involved in isoflavan phytoalexin biosynthesis in Lotus japonicus by EST mining. FEBS Lett. 2006, 580, 5666–5670.
- Kaducová, M.; Eliášová, A.; Truš, K.; Bačovčinová, M.; Sklenková, K.; Pal’ove-Balang, P. Induction of pterocarpane reductase expression and accumulation of vestitol in Lotus corniculatus on UV-B irradiation. (manuscript in preparation).
- Deavours, B.E.; Liu, C.-J.; Naoumkina, M.A.; Tang, Y.; Farang, M.A.; Sumner, L.W.; Noel, J.P.; Dixon, R.A. Functional analysis of members of the isoflavone and isoflavanone O-methyltransferase enzyme families from the model legume Medicago truncatula. Plant Mol. Biol. 2006, 62, 715–733.
- Masai, M.; Arakawa, M.; Iwaya, K.; Aoki, T.; Nakagawa, T.; Ayabe, Sh.-I.; Uchiyama, H. Discriminative phytoalexin accumulation in Lotus japonicus against symbiotic and non-symbiotic microorganisms and related chemical signals. Biosci. Biotechnol. Biochem. 2013, 77, 1773–1775.
- Ueda, H.; Sugimoto, Y. Vestitol as a chemical barrier against intrusion of parasitic plant Striga hermonthica into Lotus japonicus roots. Biosci. Biotechnol. Biochem. 2010, 74, 1662–1667.
- Lanot, A.; Morris, P. Elicitation of isoflavan phytoalexines. In Lotus japonicus Handbook; Márquez, A.J., Ed.; Springer: Dordrecht, The Netherlands, 2005; pp. 355–362.
- Alseekh, S.; Perez de Souza, L.; Benina, M.; Fernie, A.R. The style and substance of plant flavonoid decoration; towards defining both structure and function. Phytochemistry 2020, 174, 112347.
- Caputi, L.; Malnoy, M.; Goremykin, V.; Nikiforova, S.; Martens, S. A genome-wide phylogenetic reconstruction of family 1 UDPglycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012, 69, 1030–1042.
- Yin, Q.; Shen, G.; Chang, Z.; Tang, Y.; Gao, H.; Pang, Y. Involvement of tree putative glucosyltransferases from the UGT72 family in flavonol glucoside/rhamnoside biosynthesis in Lotus japonicus seeds. J. Exp. Bot. 2017, 68, 597–612.
- Modolo, L.V.; Blount, J.W.; Achnine, L.; Naoumkina, M.A.; Wang, X.Q.; Dixon, R.A. A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula. Plant Mol. Biol. 2007, 64, 499–518.
- Dhaubhadel, S.; Farhangkhoee, M.; Chapman, R. Identification and characterization of isoflavonoid specific glycosyltransferase and malonyltransferase from soybean seeds. J. Exp. Bot. 2008, 59, 981–994.




