
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | francesca Paradiso | + 2956 word(s) | 2956 | 2021-09-24 11:37:52 | | | |
| 2 | Jessie Wu | -69 word(s) | 2887 | 2021-11-22 03:12:10 | | |
Video Upload Options
From the development of self-aggregating, scaffold-free multicellular spheroids to the inclusion of scaffold systems, 3D models have progressively increased in complexity to better mimic native tissues. The inclusion of a third dimension in cancer models allows researchers to zoom out from a significant but limited cancer cell research approach to a wider investigation of the tumor microenvironment. This model can include multiple cell types and many elements from the extracellular matrix (ECM), which provides mechanical support for the tissue, mediates cell-microenvironment interactions, and plays a key role in cancer cell invasion. Both biochemical and biophysical signals from the extracellular space strongly influence cell fate, the epigenetic landscape, and gene expression. Specifically, a detailed mechanistic understanding of tumor cell-ECM interactions, especially during cancer invasion, is lacking.
1. Introduction
1.1. Mechanosensing: A Biophysical Signal Travels from the Outside to the Inside of the Cells
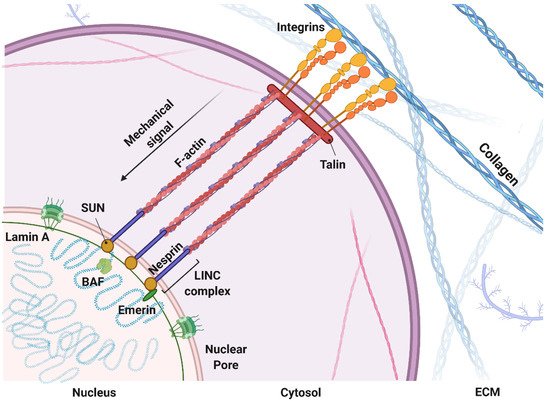
1.2. Mechanomodeling: Inclusion of Biophysical Signals in 3D Cancer Systems
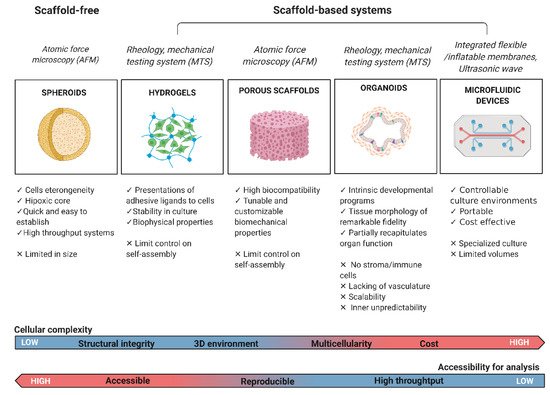
1.3. Mechanotesting: Technologies to Approach Biophysical Studies in 3D Cancer Modeling
2. Current Insight on 3D Model in Cancer
3. Future Directions
References
- Maman, S.; Witz, I.P. A history of exploring cancer in context. Nat. Rev. Cancer 2018, 18, 359–376.
- Balkwill, F.R.; Capasso, M.; Hagemann, T. The tumor microenvironment at a glance. J. Cell Sci. 2012, 125, 5591–5596.
- Bussard, K.M.; Mutkus, L.; Stumpf, K.; Gomez-Manzano, C.; Marini, F.C. Tumor-Associated stromal cells as key contributors to the tumor microenvironment. Breast Cancer Res. 2016, 18, 84.
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70.
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437.
- Tomás-Bort, E.; Kieler, M.; Sharma, S.; Candido, J.B.; Loessner, D. 3D approaches to model the tumor microenvironment of pancreatic cancer. Theranostics 2020, 10, 5074–5089.
- Chaudhuri, O.; Koshy, S.; Da Cunha, C.B.; Shin, J.-W.; Verbeke, C.S.; Allison, K.H.; Mooney, D. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat. Mater. 2014, 13, 970–978.
- Lu, P.; Takai, K.; Weaver, V.M.; Werb, Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb. Perspect. Biol. 2011, 3, a005058.
- Daley, W.P.; Peters, S.; Larsen, M. Extracellular matrix dynamics in development and regenerative medicine. J. Cell Sci. 2008, 121, 255–264.
- Romani, P.; Valcarcel-Jimenez, L.; Frezza, C.; Dupont, S. Crosstalk between mechanotransduction and metabolism. Nat. Rev. Mol. Cell Biol. 2020, 22, 22–38.
- Levental, K.; Yu, H.; Kass, L.; Lakins, J.N.; Egeblad, M.; Erler, J.; Fong, S.F.; Csiszar, K.; Giaccia, A.; Weninger, W.; et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 2009, 139, 891–906.
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120.
- Garnier, L.; Gkountidi, A.-O.; Hugues, S. Tumor-Associated lymphatic vessel features and immunomodulatory functions. Front. Immunol. 2019, 10, 720.
- Padera, T.P.; Meijer, E.; Munn, L.L. The lymphatic system in disease processes and cancer progression. Annu. Rev. Biomed. Eng. 2016, 18, 125–158.
- Mohammadi, H.; Sahai, E. Mechanisms and impact of altered tumour mechanics. Nature 2018, 20, 766–774.
- Coban, B.; Bergonzini, C.; Zweemer, A.J.M.; Danen, E.H.J. Metastasis: Crosstalk between tissue mechanics and tumour cell plasticity. Br. J. Cancer 2020, 124, 49–57.
- Wang, N. Instant integrin mechanosensing. Nat. Mater. 2017, 16, 1173–1174.
- Sun, Z.; Guo, S.S.; Fässler, R. Integrin-mediated mechanotransduction. J. Cell Biol. 2016, 215, 445–456.
- Jang, I.; Beningo, K. Integrins, CAFs and mechanical forces in the progression of cancer. Cancers 2019, 11, 721.
- Revach, O.-Y.; Grosheva, I.; Geiger, B. Biomechanical regulation of focal adhesion and invadopodia formation. J. Cell Sci. 2020, 133.
- Hirata, H.; Sokabe, M.; Lim, C.T. Molecular mechanisms underlying the force-dependent regulation of actin-to-ECM linkage at the focal adhesions. Prog. Mol. Biol. Transl. Sci. 2014, 126, 135–154.
- Kirby, T.; Lammerding, J. Emerging views of the nucleus as a cellular mechanosensor. Nature 2018, 20, 373–381.
- Fedorchak, G.R.; Kaminski, A.; Lammerding, J. Cellular mechanosensing: Getting to the nucleus of it all. Prog. Biophys. Mol. Biol. 2014, 115, 76–92.
- Enyedi, B.; Niethammer, P. A case for the nuclear membrane as a mechanotransducer. Cell. Mol. Bioeng. 2016, 9, 247–251.
- Alam, S.; Lovett, D.B.; Dickinson, R.B.; Roux, K.J.; Lele, T.P. Nuclear forces and cell mechanosensing. Prog. Mol. Biol. Transl. Sci. 2014, 126, 205–215.
- Wang, N.; Tytell, J.D.; Ingber, D.E. Mechanotransduction at a distance: Mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 2009, 10, 75–82.
- Hampoelz, B.; Azou-Gros, Y.; Fabre, R.; Markova, O.; Puech, P.-H.; Lecuit, T. Microtubule-Induced nuclear envelope fluctuations control chromatin dynamics in Drosophila embryos. Development 2011, 138, 3377–3386.
- Li, Y.; Chu, J.S.; Kurpinski, K.; Li, X.; Bautista, D.M.; Yang, L.; Sung, K.-L.P.; Li, S. Biophysical regulation of histone acetylation in mesenchymal stem cells. Biophys. J. 2011, 100, 1902–1909.
- Cho, S.; Irianto, J.; Discher, D.E. Mechanosensing by the nucleus: From pathways to scaling relationships. J. Cell Biol. 2017, 216, 305–315.
- Riehl, B.D.; Kim, E.; Bouzid, T.; Lim, J.Y. The role of microenvironmental cues and mechanical loading milieus in breast cancer cell progression and metastasis. Front. Bioeng. Biotechnol. 2021, 8, 1571.
- Alcoser, T.A.; Bordeleau, F.; Carey, S.P.; Lampi, M.C.; Kowal, D.R.; Somasegar, S.; Varma, S.; Shin, S.J.; Reinhart-King, C.A. Probing the biophysical properties of primary breast tumor-derived fibroblasts. Cell. Mol. Bioeng. 2014, 8, 76–85.
- Azadi, S.; Shadpour, M.T.; Warkiani, M.E. Characterizing the effect of substrate stiffness on the extravasation potential of breast cancer cells using a 3D microfluidic model. Biotechnol. Bioeng. 2020, 118, 823–835.
- Vitale, C.; Fedi, A.; Marrella, A.; Varani, G.; Fato, M.; Scaglione, S. 3D perfusable hydrogel recapitulating the cancer dynamic environment to In Vitro investigate metastatic colonization. Polymers 2020, 12, 2467.
- Pasini, A.; Lovecchio, J.; Cortesi, M.; Liverani, C.; Spadazzi, C.; Mercatali, L.; Ibrahim, T.; Giordano, E. Perfusion flow enhances viability and migratory phenotype in 3D-cultured breast cancer cells. Ann. Biomed. Eng. 2021, 1–11.
- Bascetin, R.; Laurent-Issartel, C.; Blanc-Fournier, C.; Vendrely, C.; Kellouche, S.; Carreiras, F.; Gallet, O.; Leroy-Dudal, J. A biomimetic model of 3D fluid extracellular macromolecular crowding microenvironment fine-tunes ovarian cancer cells dissemination phenotype. Biomaterials 2020, 269, 120610.
- Deszcz, I.; Lis-Nawara, A.; Grelewski, P.; Dragan, S.; Bar, J. Utility of direct 3D co-culture model for chondrogenic differentiation of mesenchymal stem cells on hyaluronan scaffold (Hyaff-11). Regen. Biomater. 2020, 7, 543–552.
- Gogoi, M.; Tewari, R.P. 3D printed chitosan composite scaffold for chondrocytes differentiation. Curr. Med. Imaging 2021, 17, 832–842.
- Norouzi, M.; Nazari, B.; Miller, D.W. Injectable hydrogel-based drug delivery systems for local cancer therapy. Drug Discov. Today 2016, 21, 1835–1849.
- Nii, T.; Makino, K.; Tabata, Y. A cancer invasion model combined with cancer-associated fibroblasts aggregates incorporating gelatin hydrogel microspheres containing a p53 inhibitor. Tissue Eng. Part C Methods 2019, 25, 711–720.
- Cao, H.; Lee, M.K.H.; Yang, H.; Sze, S.K.; Tan, N.S.; Tay, C.Y. Mechanoregulation of cancer-associated fibroblast phenotype in three-dimensional interpenetrating hydrogel networks. Langmuir 2018, 35, 7487–7495.
- Jensen, C.; Teng, Y. Is it time to start transitioning from 2D to 3D cell culture? Front. Mol. Biosci. 2020, 7, 33.
- Brock, E.J.; Ji, K.; Shah, S.; Mattingly, R.R.; Sloane, B.F. In Vitro models for studying invasive transitions of ductal carcinoma In Situ. J. Mammary Gland. Biol. Neoplasia 2018, 24, 1–15.
- Lintz, M.; Muñoz, A.; Reinhart-King, C.A. The mechanics of single cell and collective migration of tumor cells. J. Biomech. Eng. 2017, 139, 021005–0210059.
- Friedl, P.; Bröcker, E.B. The biology of cell locomotion within three-dimensional extracellular matrix. Cell Mol. Life Sci. 2000, 57, 41–64.
- Liu, C.; Mejia, D.L.; Chiang, B.; Luker, K.E.; Luker, G.D. Hybrid collagen alginate hydrogel as a platform for 3D tumor spheroid invasion. Acta Biomater. 2018, 75, 213–225.
- Kar, S.; Molla, M.S.; Katti, D.R.; Katti, K.S. Tissue-engineered nanoclay-based 3D in vitro breast cancer model for studying breast cancer metastasis to bone. J. Tissue Eng. Regen. Med. 2018, 13, 119–130.
- Holen, I.; Nutter, F.; Wilkinson, J.M.; Evans, C.A.; Avgoustou, P.; Ottewell, P.D. Human breast cancer bone metastasis In Vitro and In Vivo: A novel 3D model system for studies of tumour cell-bone cell interactions. Clin. Exp. Metastasis 2015, 32, 689–702.
- Devarasetty, M.; Dominijanni, A.; Herberg, S.; Shelkey, E.; Skardal, A.; Soker, S. Simulating the human colorectal cancer microenvironment in 3D tumor-stroma co-cultures In Vitro and In Vivo. Sci. Rep. 2020, 10, 9832.
- Chhetri, A.; Chittiboyina, S.; Atrian, F.; Bai, Y.; Delisi, D.A.; Rahimi, R.; Garner, J.; Efremov, Y.; Park, K.; Talhouk, R.; et al. Cell culture and coculture for oncological research in appropriate microenvironments. Curr. Protoc. Chem. Biol. 2019, 11, e65.
- Shimojo, A.A.M.; Rodrigues, I.C.P.; Perez, A.G.M.; Souto, E.M.B.; Gabriel, L.P.; Webster, T. Scaffolds for Tissue Engineering: A State-of-the-Art Review Concerning Types, Properties, Materials, Processing, and Characterization. In Racing for the Surface: Antimicrobial and Interface Tissue Engineering; Li, B., Blanco, I., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 647–676.
- Qiao, H.; Tang, T. Engineering 3D approaches to model the dynamic microenvironments of cancer bone metastasis. Bone Res. 2018, 6, 3.
- Díaz, E.C.G.; Sinha, S.; Avedian, R.S.; Yang, F. Tissue-engineered 3D models for elucidating primary and metastatic bone cancer progression. Acta Biomater. 2019, 99, 18–32.
- Liaw, C.Y.; Ji, S.; Guvendiren, M. Engineering 3D hydrogels for personalized In Vitro human tissue models. Adv. Healthc. Mater. 2018, 7, 1701165.
- García-Gareta, E.; Abduldaiem, Y.; Sawadkar, P.; Kyriakidis, C.; Lali, F.; Greco, K.V. Decellularised scaffolds: Just a framework? Current knowledge and future directions. J. Tissue Eng. 2020, 11, 2041731420942903.
- Hoshiba, T. Decellularized extracellular matrix for cancer research. Materials 2019, 12, 1311.
- Jin, Q.; Liu, G.; Li, S.; Yuan, H.; Yun, Z.; Zhang, W.; Zhang, S.; Dai, Y.; Ma, Y. Decellularized breast matrix as bioactive microenvironment for In Vitro three-dimensional cancer culture. J. Cell. Physiol. 2018, 234, 3425–3435.
- Mohammadkarim, A.; Mokhtari-Dizaji, M.; Kazemian, A.; Saberi, H.; Khani, M.M.; Bakhsh, M. The mechanical characteristics of human endothelial cells in response to single ionizing radiation doses by using micropipette aspiration technique. Mol. Cell. Biomech. 2019, 16, 275–287.
- González-Bermúdez, B.; Guinea, G.V.; Plaza, G.R. Advances in micropipette aspiration: Applications in cell biomechanics, models, and extended studies. Biophys. J. 2019, 116, 587–594.
- Hogan, B.; Babataheri, A.; Hwang, Y.; Barakat, A.I.; Husson, J. Characterizing cell adhesion by using micropipette aspiration. Biophys. J. 2015, 109, 209–219.
- Yousafzai, M.S.; Coceano, G.; Bonin, S.; Niemela, J.; Scoles, G.; Cojoc, D. Investigating the effect of cell substrate on cancer cell stiffness by optical tweezers. J. Biomech. 2017, 60, 266–269.
- Huang, H.; Dai, C.; Shen, H.; Gu, M.; Wang, Y.; Liu, J.; Chen, L.; Sun, L. Recent advances on the model, measurement technique, and application of single cell mechanics. Int. J. Mol. Sci. 2020, 21, 6248.
- Kim, H.; Yamagishi, A.; Imaizumi, M.; Onomura, Y.; Nagasaki, A.; Miyagi, Y.; Okada, T.; Nakamura, C. Quantitative measurements of intercellular adhesion between a macrophage and cancer cells using a cup-attached AFM chip. Colloids Surf. B Biointerfaces 2017, 155, 366–372.
- Hayashi, K.; Iwata, M. Stiffness of cancer cells measured with an AFM indentation method. J. Mech. Behav. Biomed. Mater. 2015, 49, 105–111.
- Luo, Q.; Kuang, D.; Zhang, B.; Song, G. Cell stiffness determined by atomic force microscopy and its correlation with cell motility. Biochim. Biophys. Acta-Gen. Subj. 2016, 1860, 1953–1960.
- Xu, W.; Mezencev, R.; Kim, B.; Wang, L.; McDonald, J.F.; Sulchek, T. Cell stiffness is a biomarker of the metastatic potential of ovarian cancer cells. PLoS ONE 2012, 7, e46609.
- Li, M.; Xi, N.; Wang, Y.-C.; Liu, L.-Q. Atomic force microscopy for revealing micro/nanoscale mechanics in tumor metastasis: From single cells to microenvironmental cues. Acta Pharmacol. Sin. 2020, 42, 323–339.
- Taubenberger, A.V.; Girardo, S.; Träber, N.; Fischer-Friedrich, E.; Kräter, M.; Wagner, K.; Kurth, T.; Richter, I.; Haller, B.; Binner, M.; et al. 3D Microenvironment stiffness regulates tumor spheroid growth and mechanics via p21 and ROCK. Adv. Biosyst. 2019, 3, 1900128.
- Glentis, A.; Oertle, P.; Mariani, P.; Chikina, A.; Marjou, F.E.; Attieh, Y.; Zaccarini, F.; Lae, M.; Loew, D.; Dingli, F.; et al. Cancer-associated fibroblasts induce metalloprotease-independent cancer cell invasion of the basement membrane. Nat. Commun. 2017, 8, 924.
- Picout, D.R.; Ross-Murphy, S.B. Rheology of biopolymer solutions and gels. Sci. World J. 2003, 3, 105–121.
- Janmey, P.A.; Schliwa, M. Rheology. Curr. Biol. 2008, 18, R639–R641.
- Mathieu, S.; Manneville, J.-B. Intracellular mechanics: Connecting rheology and mechanotransduction. Curr. Opin. Cell Biol. 2018, 56, 34–44.
- Andrikakou, P.; Vickraman, K.; Arora, H. On the behaviour of lung tissue under tension and compression. Sci. Rep. 2016, 6, 36642.
- Park, S.; Ateshian, G.A. Dynamic response of immature bovine articular cartilage in tension and compression, and nonlinear viscoelastic modeling of the tensile response. J. Biomech. Eng. 2006, 128, 623–630.
- Frank, E.H.; Jin, M.; Loening, A.; Levenston, M.; Grodzinsky, A.J. A versatile shear and compression apparatus for mechanical stimulation of tissue culture explants. J. Biomech. 2000, 33, 1523–1527.
- Deptuła, P.; Łysik, D.; Pogoda, K.; Cieśluk, M.; Namiot, A.; Mystkowska, J.; Król, G.; Głuszek, S.; Janmey, P.A.; Bucki, R. Tissue rheology as a possible complementary procedure to advance histological diagnosis of colon cancer. ACS Biomater. Sci. Eng. 2020, 6, 5620–5631.
- Sutherland, R.M.; McCredie, J.A.; Inch, W.R. Growth of multicell spheroids in tissue culture as a model of nodular carcinomas2. J. Natl. Cancer Inst. 1971, 46, 113–120.
- Alemany-Ribes, M.; Semino, C.E. Bioengineering 3D environments for cancer models. Adv. Drug Deliv. Rev. 2014, 79–80, 40–49.
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011, 21, 745–754.
- Roy, V.; Magne, B.; Vaillancourt-Audet, M.; Blais, M.; Chabaud, S.; Grammond, E.; Piquet, L.; Fradette, J.; Laverdière, I.; Moulin, V.J.; et al. Human organ-specific 3D cancer models produced by the stromal self-assembly method of tissue engineering for the study of solid tumors. BioMed Res. Int. 2020, 2020, 6051210.
- Pati, F.; Gantelius, J.; Svahn, H.A. 3D bioprinting of tissue/organ models. Angew. Chem. Int. Ed. 2016, 55, 4650–4665.
- Alonso-Alconada, L.; de la Fuente, A.; Santacana, M.; Ferreiros, A.; Lopez-Lopez, R.; Matias-Guiu, X.; Abal, M. Biomimetic device and foreign body reaction cooperate for efficient tumour cell capture in murine advanced ovarian cancer. Dis. Model. Mech. 2020, 13, dmm043653.
- Fong, E.L.S.; Toh, T.B.; Yu, H.; Chow, E. 3D culture as a clinically relevant model for personalized medicine. SLAS Technol. Transl. Life Sci. Innov. 2017, 22, 245–253.
- Sensi, F.; D’Angelo, E.; Piccoli, M.; Pavan, P.; Mastrotto, F.; Caliceti, P.; Biccari, A.; Corallo, D.; Urbani, L.; Fassan, M.; et al. Recellularized colorectal cancer patient-derived scaffolds as In Vitro pre-clinical 3D model for drug screening. Cancers 2020, 12, 681.
- D’Angelo, E.; Natarajan, D.; Sensi, F.; Ajayi, O.; Fassan, M.; Mammano, E.; Pilati, P.; Pavan, P.; Bresolin, S.; Preziosi, M.; et al. Patient-Derived scaffolds of colorectal cancer metastases as an organotypic 3D model of the liver metastatic microenvironment. Cancers 2020, 12, 364.
- Piccoli, M.; D’Angelo, E.; Crotti, S.; Sensi, F.; Urbani, L.; Maghin, E.; Burns, A.; De Coppi, P.; Fassan, M.; Rugge, M.; et al. Decellularized colorectal cancer matrix as bioactive microenvironment for in vitro 3D cancer research. J. Cell. Physiol. 2017, 233, 5937–5948.
- Agarwal, T.; Rustagi, A.; Das, J.; Maiti, T.K. PAMAM dendrimer grafted cellulose paper scaffolds as a novel in vitro 3D liver model for drug screening applications. Colloids Surf. B Biointerfaces 2018, 172, 346–354.
- Candini, O.; Grisendi, G.; Foppiani, E.M.; Brogli, M.; Aramini, B.; Masciale, V.; Spano, C.; Petrachi, T.; Veronesi, E.; Conte, P.; et al. A novel 3D In Vitro platform for pre-clinical investigations in drug testing, gene therapy, and immuno-oncology. Sci. Rep. 2019, 9, 7154.
- Brancato, V.; Oliveira, J.M.; Correlo, V.M.; Reis, R.L.; Kundu, S.C. Could 3D models of cancer enhance drug screening? Biomaterials 2020, 232, 119744.
- Booij, T.H.; Price, L.S.; Danen, E.H.J. 3D cell-based assays for drug screens: Challenges in imaging, image analysis, and high-content analysis. SLAS Discov. Adv. Life Sci. R&D 2019, 24, 615–627.
- Augustine, R.; Kalva, S.N.; Ahmad, R.; Zahid, A.A.; Hasan, S.; Nayeem, A.; McClements, L.; Hasan, A. 3D Bioprinted cancer models: Revolutionizing personalized cancer therapy. Transl. Oncol. 2021, 14, 101015.
- Van Zundert, I.; Fortuni, B.; Rocha, S. From 2D to 3D cancer cell models—The enigmas of drug delivery research. Nanomaterials 2020, 10, 2236.
- Nii, T.; Makino, K.; Tabata, Y. Three-Dimensional culture system of cancer cells combined with biomaterials for drug screening. Cancers 2020, 12, 2754.
- Yang, M.; Li, J.; Gu, P.; Fan, X. The application of nanoparticles in cancer immunotherapy: Targeting tumor microenvironment. Bioact. Mater. 2020, 6, 1973–1987.
- Nii, T.; Katayama, Y. Biomaterial-Assisted regenerative medicine. Int. J. Mol. Sci. 2021, 22, 8657.




