
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Leandro Rodríguez-Viera | + 3503 word(s) | 3503 | 2021-10-09 04:56:02 | | | |
| 2 | Catherine Yang | Meta information modification | 3503 | 2021-10-21 03:03:04 | | |
Video Upload Options
Decapod crustaceans are a very diverse group and have evolved to adapt to a broad variety of diets. However, α-amylases have been more thoroughly studies in herbivore and omnivore species, both from an evolutionary/ecological and applied (i.e., aquaculture) point of view, while information on α-amylases from carnivorous species is scarce. Diverse studies revealed that enzyme sequences and overall architecture is highly conserved among decapods.
1. Introduction
Decapod crustaceans diverged in the Late Ordovician and most lineages diverged in the Triassic–Jurassic [1]. Since then, this group of animals has experienced a great diversification, and today over 15,000 living species populate marine, freshwater, and semi-terrestrial environments [2]. This ecological success relies, to a great extent, in the capacity of the different groups to adapt to a broad variety of diets. Indeed, decapods exhibit a wide variation in feeding habits, which includes herbivores, carnivores, scavengers, deposit feeders, filter feeders, and opportunistic omnivores [3][4]. In addition, their wide geographic distribution implies that digestion of such a variety of foods occurs over an extensive range of environmental conditions (e.g., temperature, salinity, etc.). After ingestion, digestive enzymes are responsible for the hydrolysis of complex dietary components into assimilable nutrients and accordingly, digestive enzymes harbored by decapods have been studied, although less deeply than in other arthropods such as insects [5].
The digestive enzymes of omnivore crabs [6][7][8][9][10][11] and shrimps [12][13] have been studied from an evolutionary perspective because differences between plants and animals force trade-offs in the traits required to use these feeds simultaneously [14]. Likewise, digestive enzymes adaptation to a vegetarian diet has been studied in different species [15] as at least 31 lineages of marine, freshwater, and terrestrial crustaceans have independently overcome the challenge of consuming plant material [16]. In the case of penaeid shrimps, digestive enzymes studies, and the direct relationship between digestive enzymes and feed utilization, have also been speeded up due to their economic importance in aquaculture worldwide [17]. Conversely, carnivorous species have been historically less studied. However, information has been produced during the last decade on the digestive biochemistry of a carnivorous spiny lobster [18][19][20][21][22][23][24][25], shedding light on aspects such as isoenzyme richness, molecular, and biochemical differences among isoforms, molecular evolution, and regulatory mechanisms.
Crustaceans with particular feeding habits exhibit distinctive digestive enzymes, such as cellulase and hemicellulase in those that feed on leaves [3][26][27][28], or laminarinase in those consuming brown and green phytoplankton and algae [26]. However, all species share main digestive enzymes such as proteases (trypsin, chymotrypsin, etc.), lipases, and α-amylases (α-1,4-alpha-D-glucan glucanohydrolase, EC 3.2.1.1; henceforth named α-amylases). Protein and lipids are well known to be key nutrients for crustaceans metabolism [29][30] while the role of dietary carbohydrates is not that clear and rather variable among species. Even when dietary carbohydrate cannot be efficiently used by aquatic animals [31], carbohydrates are essential and thus included in artificial feeds at 20% to 30% [32][33], although higher carbohydrate intake can lead to slow growth, low immunity, and high mortality rates [31][32]. Among carbohydrases, α-amylase is responsible for the hydrolysis of starch and glycogen, but remained poorly studied in carnivorous decapods until recently [18][19][23][24][25][34]. The new information provided by these recent studies now allows drawing a more comprehensive view of the role of α-amylases across decapods crustaceans.
2. Molecular Features
The presence of several α-amylase gene copies may be advantageous for more enzyme production, for fine developmental and tissue-specific expression, for broadening pH and substrate range, or for overcoming the natural defenses of plants if they are included in diet [5]. Molecular information on the α-amylase gene in the crustacean decapod is restricted to few species. In the omnivorous shrimp Litopenaeus vannamei , three α-amylase genes have been characterized, with nine introns located at the same positions but presenting no similarity among genes [35]. However, an RNA-seq study found 16 unigenes for α-amylase in this species [36]. Within the Panama natural population, 35 different alleles occur at this locus [35]. In the shrimp Palaemonetes varians, population studies found four co-dominant alleles, while some populations only exhibit two of them [37]. In contrast, a single and intron-less gene occurs in the carnivorous lobster P. argus [24]. The number of α-amylase genes is also variable in non-decapod crustaceans. For instance, six copies of the α-amylase gene occur in the detritivore isopod Asellus aquaticus [38], which eats on leaf material in freshwater environments [39], while two copies occur in another detritivore isopod, Sphaeroma serratum [38], which fed on detritus from marine algae or terrestrial plants [39], although its fatty acid signature suggested that animal material is also included in its natural diet. In other arthropods this issue has been studied more thoroughly. In insects, the copy number varies from only 1 (e.g, in honeybees) to more than 12 (in some mosquitoes) [5]. Among them, α-amylase genes have been more thoroughly studied in Drosophila , and the number of gene copies within this single genera varies from 1 to 6 [38].
The spiny lobster ( Panulirus argus ) gene encodes a single transcript (PaAmy, GenBank accession no. LK937698) of 1830 bp, with a short 5′ untranslated region of 23 bp, a long 3’ untranslated region of 268 bp, and a 1539 bp ORF. Before the poly A tail, two sites of alternative polyadenylation were found at 108 bp and 139 bp downstream the stop codon. The lobster transcript exhibited high identity with α-amylases cDNAs from other decapods such as L. vannamei (79%) and Penaeus japonicus (78%) α-amylase, but also high ( >60%) with α-amylases from phylogenetically distant groups such as humans ( Table 1 ).
Table 1. Conservation (i.e., identity) of the lobster Panulirus argus α-amylase cDNA sequence (GenBank accession no. LK937698, 1830 bp long) with respect to other α-amylases from decapod crustaceans and humans.
| Group | Species | Accession No. Genbank | Identity (%) | Nucleotides (pb) |
|---|---|---|---|---|
| Brachyurans | Eriocheir sinensis | KU301756.1 | 75.6 | 1663 |
| Helice tientsinensis | MN964184.1 | 75.39 | 1527 | |
| Neohelice granulata | KU531567.1 | 75.04 | 1637 | |
| Macrophthalmus pacificus | MN964194.1 | 74.34 | 1533 | |
| Gelasimus borealis | MN964240.1 | 76.19 | 1533 | |
| Metopograpsus quadridentatus | MN964203.1 | 76.25 | 1533 | |
| Parasesarma pictum | MN964222.1 | 76.22 | 1533 | |
| Parasesarma affine | MN964213.1 | 76.09 | 1533 | |
| Chiromantes dehaani | MN964164.1 | 75.36 | 1533 | |
| Sesarmops sinensis | MN964231.1 | 75.21 | 1215 | |
| Calappa philargius | MN964146.1 | 69.98 | 1533 | |
| Charybdis japonica | MN964155.1 | 76.32 | 1533 | |
| Scylla olivacea | GDRN01093055.1 | 75.51 | 1715 | |
| Portunus trituberculatus | MN964137.1 | 74.37 | 1533 | |
| Penaeids | Marsupenaeus japonicus | KJ147432.1 | 77.95 | 1651 |
| Penaeus monodon | KU308415.1 | 66.34 | 2465 | |
| Litopenaeus vannamei | KM077131.1 | 66.17 | 2358 | |
| Carideans | Crangon crangon | MH055762.1 | 66.43 | 2175 |
| Macrobrachium rosenbergii | KM886337.1 | 67.6 | 2282 | |
| Astacids | Astacus leptodactylus | KF954216 | 65.69 | 2250 |
| Homarus americanus | XM_042364069 | 67.45 | 2434 | |
| Procambarus clarkii | MF688642.1 | 67.17 | 2138 | |
| Human | Homo sapiens | M24895.1 | 66.85 | 1612 |
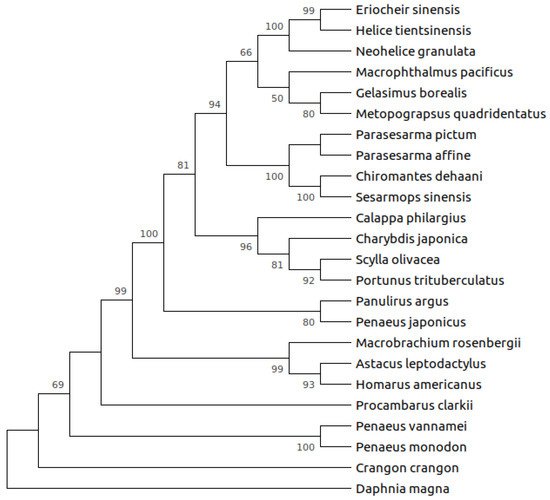
There is not a clear and complete picture of α-amylase evolution within decapods crustaceans. A previous phylogenetic analysis including α-amylases from shrimps and lobsters, and those of insects, fishes, amphibians, birds, and mammals, retrieved the expected topology resembling phylogenetic relationships among groups [24]. Within the well-supported Arthropoda clade, crustacean’s α-amylases appeared as a monophyletic group [24]. However, more α-amylase sequences are now available ( Table 1 ), and this allows having a wider view on their sequence evolution, although there are more sequences for crabs than for other groups. Evolutionary analyses of crab’s α-amylases found evidence of positive selection in the enzyme of herbivore crabs, whereas not in omnivore or carnivore species [11]. Nevertheless, a wider analysis, including α-amylases from major groups of decapods crustaceans revealed that while most crab α-amylases appear as a monophyletic group which further diversify, α-amylases from phylogenetically distant groups such as shrimps and lobsters clustered together according to their feeding habits (i.e., carnivores or omnivores) ( Figure 1 ), suggesting that convergent evolution might have occurred among distant lineages with feeding habits as a selection force. Indeed, ongoing analyses at our laboratory revealed that positive selection also occurred at common sites in omnivore species from distant groups such as shrimps and crayfishes.
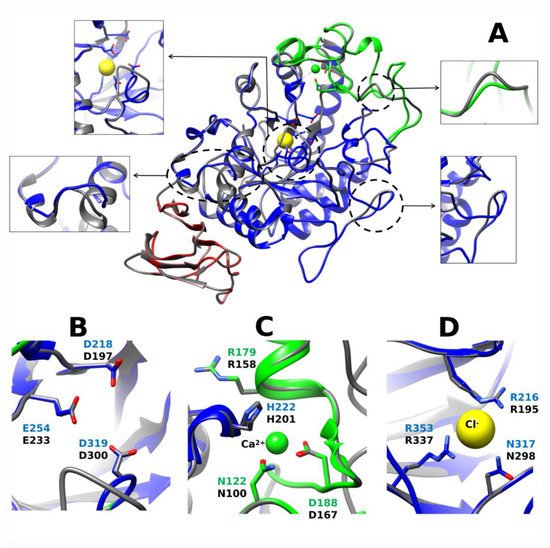
Alpha-amylase enzymes in decapod crustaceans have estimated molecular weights between 26 and 75 kDa. Estimates differ depending on whether they come from electrophoresis mobility or from cDNA sequences. For example, the molecular weight for the lobster P. argus α-amylase was estimated to be around 44–47 kDa [18] by electrophoresis whereas 55.5 kDa from its transcript sequence [24]. Few protein sequences for α-amylase of decapod crustaceans are available. The lobster transcript encodes a protein with 513 amino acids, including a highly hydrophobic signal peptide of 21 amino acids, a potential cleavage site for the signal peptide between Ala21 and Gln22, and predicted molecular mass and isoelectric point for the mature enzyme of 55.5 kDa and 4.93, respectively. The comparison of amino acid sequence of lobster enzyme and other α-amylases showed a high similarity in conserved regions I to VI, but region VII was not identified. The region VII is known to be less conserved among the family [40]. A model for this α-amylase was developed and deposited at the Protein Model Data Base ( http://bioinformatics.cineca.it/PMDB/main.php ), accessed on 11 June 2021 under PMDB id: PM0079556. The enzyme has the typical 3D structure of α-amylase enzymes. It is formed by three domains A, B, C. Domain A is a (β/α)8-barrel, B is a loop between the β3 strand and α3 helix of A, and C is the C-terminal extension. PaAmy has the active site cleft between domains A and B, with a triad of catalytic residues (Asp218, Glu255 and Asp319). It contains a calcium-binding site (Asn122, Arg179, Asp194, and His222), a chloride-binding site (Arg216, Asn317, and Arg353), and several cysteines residues ( Figure 2 A). Ten cysteines residues were observed in the lobster α-amylase, as occur in α-amylases from other arthropods [12][41]. Eight of these cysteines are also conserved in vertebrate α-amylases [42]. The additional two residues in crustaceans and other invertebrates enable a fifth disulfide bridge, and may be related with differences in activity during temperature adaptation [12]. In general, overall architecture of the α-amylase is highly conserved, even when compared with the human enzyme ( Figure 2 A), although some differences occur in superficial loops which effects on enzyme function are unknown. These effects, if any, may be related with extended interactions with large substrates. Given that these regions are subjected to less evolutionary constrains, their analysis in carnivore, omnivore, and herbivore species may shed light on their evolution across decapod crustaceans, but this examination have been not yet produced. Notably, the geometry of key residues for α-amylase function such as the catalytic triad, and the binding sites for calcium and chloride are highly similar in the lobster and the human enzyme ( Figure 2 B–D).
3. Biochemical Features
The α-amylase activity in the marine crab Maguimithrax spinosissimus is poorly affected by NaCl [43], although it has been reported that NaCl influences the α-amylase activity in marine shrimps [44], crabs [10], and lobsters [24][45]. For instance, in estuarine amphipod Gammarus palustris , activation occurred at low chloride concentrations, achieving 90% of the maximum activity at 8 mM NaCl, but no inhibition occurred at higher concentrations [46]. In the estuarine shrimp Farfantepenaeus californiensis , α-amylase activity is highest at a low salt concentration (i.e., 0.01 M NaCl), and it is also poorly affected by high salt concentration, retaining 50% of its activity at 3 M NaCl [44]. Likewise, while α-amylase activity in the euryhaline burrowing crab Neohelice granulata is maximal in the wide range of 0.5–1.5 M, it is maintained at high NaCl concentrations (up to 4 M), retaining 30% of initial activity [10]. On the other hand, while in larvae of the marine lobster Homarus americanus α-amylase activity does not significantly vary over the range 0.05–0.2 M NaCl [47], in adult lobsters, activation of the enzyme is highest at 0.1 M NaCl [45], and in the marine spiny lobster P. argus at 0.3 M NaCl [24]. In summary, differences occur among the crustacean α-amylases in their response to salt concentration, probably reflecting habitat features and/or evolutionary relationships.
At least one calcium binding site occurs in α-amylases [48][49]. Studies in the crabs Carcinus maenas [50] , N. granulata [10], and M. spinosissimus [43]; in the spiny lobster P. argus [24]; in the crayfish Cherax quadricarinatus [51]; in three species of penaeid shrimps [13]; and in other invertebrates [52][53][54] have showed enhancements in α-amylase activity when CaCl 2 concentration increases up to a maximum and then decreases. However, exceptions occurred, as in the lobster H. americanus , where no effect of calcium was reported [45], while in the crab Portunus segnis , only minor effect of calcium was reported [55]. Calcium binding sites are important structural features of amylase enzymes, and it is well known that this ion is important for the activity and stability of α-amylases. However, the stability effects demonstrated by calcium in decapod amylases are not well understood, and this issue has never been evaluated in many species.
In general, the thermal stability of α-amylases is relatively low above 30–37 °C in shrimps [13] and also in some non-decapod species [56]. Although the α-amylase of the tropical king crab M. spinosissimus was stable at a high temperature (>50 °C) [43], α-amylase activity from the tropical lobster P. argus is compromised above 30 °C [24], as in other crustaceans. Yet, in the crab P. segnis , which is tolerant of a wide range of temperatures from 13 °C to 30 °C, the enzyme is also highly stable at 50 °C [55]. Less variation has been observed in optimal temperature, i.e., lobster P. argus , 50 °C [24]; shrimp F. californiensis, 30–40 °C [44]; and different species of crab, e.g., Scylla serrata , 50 °C [57], N. granulata, 30–40 °C [10]; and P. segnis 50 °C [55]. In summary, the relationship between stability and habitat temperature is not clear, probably because this feature mostly depends on the conserved architecture of the enzyme among crustaceans.
With the few studies available and the disparity in methodologies employed for the kinetic determination of α-amylases in decapods, results do not allow the drawing of clear relationships between catalytic activities and other characteristics of animals such as taxonomy or feeding habits. However, the detailed study of Van Wormhoudt and colleagues [58] provided important information in this regard. In that study, shrimps and crabs showed the highest activity among 40 species analyzed, while comparatively low activity in one carnivorous spiny lobster species. Yet, the study reported very few differences in the specific activities of the pure enzymes, suggesting that the catalytic features of α-amylases from crustacean decapods might be similar [58]. Thus, differences in activity among groups or species might be more related to the amount of enzyme synthesized and/or secreted into the digestive tract. Indeed, the α-amylase content of digestive gland of the carnivore crab C. maenas and of the carnivore-scavenger Pagurus bernhardus is about 0.1% of total proteins, whereas it was 1% in the omnivores L. vannamei and Procambarus clarkii [58]. However, little information is available on the regulation of transcription, synthesis, and secretion of α-amylases at the molecular level in crustaceans (See Section 6 ).
4. Alpha-Amylase Regulation
Variations in digestive enzyme activities reflect the maturation of the digestive system at early stages and later, changing physiological requirements as animals grow. Often, a clear relationship with shift in diet composition can be observed, while in other cases, contrasting results have been reported. In general, phytotrophic larval stages show an apparent predominance of trypsin content, while in carnivorous larvae a higher ratio of α-amylase to protease is observed [59]. However, variation occurs. For example, α-amylase activity is extremely low during carnivorous early larval stages of M. rosenbergii while increased sharply when the animal develop into an omnivore juvenile [60]. In the spider crab Maja brachydactyla, α-amylase showed a continuous enhancement of total activity through development, and zymograms revealed that α-amylase-active bands increased in number and intensity as development advanced [7]. Likewise, α-amylase activity in the predator larva of the lobster H. americanus increased slightly at the time of hatching and also during larval Stages I and II, achieving maximal activity among Stage V juveniles [47]. Conversely, in another crab, S. serrata, α-amylase activity enhanced through first stages of developments (i.e., zoea) but then gradually declined at more advanced stages [61], as also occurred in the crayfish P. clarkii [62]. Early shrimp larvae fed on phytoplankton and gradually incorporate zooplankton in their diets [63]. In the shrimps Penaeus setiferus and P. indicus , peak activities for all enzymes occurred during late zoeal or early mysis larval stages and later, α-amylase activity significantly increased during postlarval development [64][65]. In L. vannameii , 9 out of 16 unigenes enhanced their expression from nauplius to zoea contributing to a significant increase in activity [36]. However, contrasting results have been obtained in juveniles sharing similar feeding habits. For instance, the α-amylase importance in digestion seems to decrease as the omnivore anomuran crab Aegla uruguayana juveniles grown, while in the omnivore crayfish Macrobrachium borellii this trend is not evident [66]. In other omnivore crayfish, such as the redclaw C. quadricarinatus, α-amylase activity remains relatively constant in early juveniles but shows a great increase in larger animals [67].
In the carnivore lobster P. argus we found no trends in the relationship between specific α-amylase activity and size (in a range from 6 to 20 mm carapace length, i.e., from first post-pueruli to first juvenile stages) [18]. However, in juveniles and adults, there is a significant positive relationship between specific α-amylase activity and lobster size, suggesting that the capacity for carbohydrate digestion increases as lobsters grow [19] and fed on bigger prey items probably with a higher content of glycogen. Indeed, multivariate analysis suggested that in P. argus digestive enzyme activities appear to be strongly influenced by changes in diet [19]. Conversely to that found in P. argus , small Jasus edwardsii exhibit higher α-amylase activity than large specimens [34].
The ability of organisms to adapt to the characteristics of the diet to cover the requirements of certain nutrients has been documented in a wide variety of species, including crustaceans. This ability relies largely on variations in the activity levels of digestive enzymes. A positive correlation of α-amylase activity and dietary carbohydrates has been reported in very distant groups such as insects [68][69], mollusks [70], fish [71][72], dogs [73], and humans [74], and also relates positively with the amount of transporters necessary for their absorption at intestinal level [75]. In general, high amylolytic activity in herbivorous and omnivorous is accepted to result from adaptation to low energy food and low assimilation efficiency or as an adaptation to large amounts of dietary starch [58][76]. An early study comparing digestive enzymes of crustaceans with different feeding habits suggested that omnivores have more α-amylase activity than carnivorous species [76]. Much later, the most comprehensive assessment of α-amylase activity in crustaceans included 40 different species and confirmed that omnivorous crustaceans such as shrimps, crabs, and crayfish have relatively high α-amylase activity with respect to carnivorous species [58]. Other studies also reported that omnivorous crab species present high α-amylase activities [26]. In agreement, in a comparison among decapods with different feeding habits, the highest α-amylase to protease ratio was observed in adults of the omnivore shrimp Macrobrachium australiense and the lowest in mostly carnivores crabs Portunus pelagicus and S. serrata [77]. Also in this line, some herbivore crayfish exhibits higher α-amylase activity than omnivore shrimps [78]. However, few contradictory results have also been obtained. Alpha-amylase activity in adults of the omnivore shrimp P. indicus is higher than in other omnivore shrimp, L. vannamei , especially at high temperatures [79], suggesting a role of environmental temperature on this activity. Likewise, the association of α-amylase activity and diet was not clear in four land crabs species with detritivorous or omnivorous feeding habits [80]. In this regard, it is important to remark that several factors converge for the adaptation to a particular trophic level such as live history, metabolic rate, behavior, and other features of the digestive processes including food intake, mechanical digestion, retention time, and assimilation efficiency [27]. Moreover, digestive enzymes other than α-amylase often have a major role in carbohydrate digestion [27]. This is the case of enzymes that digest cellulose (endo-ß-1,4-glucanase, cellobiohydrolase, ß-1,4-glucosidase) and hemicellulose (laminarinase, lichenase, xylanase) in herbivore species such as land crabs, coincident with the higher level of these carbohydrates in their diets with respect to starch [27].
Moreover, α-amylase activity has been regularly reported in carnivorous crustaceans [18][26][34][45]. The relatively high α-amylase activity in spiny lobsters seems to contradict their limited metabolic use of carbohydrates [23][25][81], evident by the reduced activity of enzymes involved in both glycolysis and glycogen synthesis [23][25], although a recent study revealed that carbohydrate was the predominant energy substrate in 3-day fasted lobsters if previously fed a low (i.e., 40%) protein diet [82]. Yet, carbohydrates continue having a less important role as energy substrate after feeding, and even in fasted lobsters, if previously ingested a protein rich (i.e., 50%) diet [82]. Likewise, the high α-amylase activity in carnivorous larvae of the spider crab Hyas araneus does not correspond to the low carbohydrate content in its food and this was suggested to be a phylogenetic remnant from ancestor species with partly herbivorous larvae [83]. Interestingly, results in four closely related prickleback fishes showed that activity of α-amylase follows a pattern influenced more by phylogeny than by diet in these fishes [84] suggesting that this could be a common pattern.
References
- Wolfe, J.M.; Breinholt, J.W.; Crandall, K.A.; Lemmon, A.R.; Lemmon, E.M.; Timm, L.E.; Siddall, M.E.; Bracken-Grissom, H.D. A phylogenomic framework, evolutionary timeline and genomic resources for comparative studies of decapod crustaceans. Proc. Biol. Sci. 2019, 286, 20190079.
- De Grave, S.; Pentcheff, N.D.; Ahyong, S.T.; Chan, T.Y.; Crandall, K.A.; Dworschak, P.C.; Felder, D.L.; Feldmann, R.M.; Fransen, C.H.J.M.; Goulding, L.Y.D.; et al. A classification of living and fossil genera of decapod crustaceans. Raffles Bull. Zool. 2009, 21, 1–109.
- Lancia, J.P.; Fernandez-Gimenez, A.V.; Bas, C.; Spivak, E. Adaptative differences in digestive enzyme activity in the crab Neohelice granulate in relation to sex and habitat. J. Crustac. Biol. 2012, 32, 940–948.
- Davie, P.; Guinot, J.F.; Peter, D.; Ng, K.L. Systematics and classification of Brachyura. Treatise on zoology-anatomy, taxonomy, biology. Crustacea 2015, 9, 1049–1130.
- Da Lage, J.L. The amylases of insects. Inter. J. Insect Sci. 2018, 10, 1179543318804783.
- Brun, G.; Wojtowicz, M. A comparative study of the digestive enzymes in the hepatopancreas of Jonah crab (Cancer borealis) and rock crab (Cancer irroratus). Comp. Biochem. Physiol. B 1976, 53, 387–391.
- Andrés, M.; Gisbert, E.; Díaz, M.; Moyano, F.J.; Estévez, A.; Rotllant, G. Ontogenetic changes in digestive enzymatic capacities of the spider crab, Maja brachydactyla (Decapoda: Majidae). J. Exp. Mar. Biol. Ecol. 2010, 389, 75–84.
- Abol-Munafi, A.B.; Pilus, N.; Amin, R.M.; Azra, M.N.; Ikhwanuddin, M. Digestive enzyme profiles from foregut contents of blue swimming crab, Portunus pelagicus from Straits of Johor, Malaysia. J. Assoc. Arab Univ. Basic Appl. Sci. 2017, 24, 120–125.
- Asaro, A.; Martos-Sitcha, J.A.; Martínez-Rodríguez, G.; Mancera, J.M.; López-Mañanes, A.A. In silico analysis and effects of environmental salinity in the expression and activity of digestive α-amylase and trypsins from the euryhaline crab Neohelice granulata. Can. J. Zool. 2018, 96, 127–139.
- Asaro, A.; Paggi, R.A.; De Castro, R.; Lopez-Mañanes, A.A. Amylase in the hepatopancreas of a euryhaline burrowing crab: Characteristics and modulation. Turk. J. Zool. 2017, 41, 443–453.
- Wang, Z.; Tang, D.; Huayun, G.; Chenchen, S.; Wu, L.; Yaqi, L. Evolution of digestive enzyme genes associated with dietary diversity of crabs. Genetica 2020, 148, 87–99.
- Van Wormhoudt, A.; Sellos, D. Cloning and sequencmg analysis of three cDNAs in the shrimp Pertaeus vannamei: Evolutionary aspects. (Crustacea Decapoda). J. Mol. Evol. 1996, 42, 543–551.
- Castro, P.F.; Freitas, A.C.V.; Santana, W.M.; Costa, H.M.S.; Carvalho, L.B.; Bezerra, R.S. Comparative study of amylases from the midgut gland of three species of penaeid shrimp. J. Crust. Biol. 2012, 32, 607–613.
- Roitberg, B.D.; Gillespie, D.R.; Quiring, D.M.; Alma, C.R.; Jenner, W.H.; Perry, J.; Peterson, J.H.; Salomon, M.; VanLaerhoven, S. The cost of being an omnivore: Mandible wear from plant feeding in a true bug. Naturwissenschaften 2005, 92, 431–434.
- Jormalainen, V. Grazers of macroalgae and higer plants. In Natural History of the Crustacea—Lifestyles and Feeding Biology; Thiel, M., Watling, L., Eds.; Oxford University Press: Oxford, UK, 2015; 584p.
- Poore, A.G.B.; Ahyong, S.T.; Lowry, J.K.; Sotka, E.E. Plant feeding promotes diversification in the crustacea. Proc. Natl. Acad. Sci. USA 2017, 114, 8829–8834.
- FAO. The State of World Fisheries and Aquaculture 2018-Meeting the Sustainable Development Goals; Food and Agriculture Organization of the United Nations: Rome, Italy, 2018; p. 227.
- Perera, E.; Moyano, F.J.; Díaz, M.; Perdomo-Morales, R.; Montero-Alejo, V.; Alonso, E.; Carrillo, O.; Galich, G.S. Polymorphism and partial characterization of digestive enzymes in the spiny lobster Panulirus argus. Comp. Biochem. Physiol. B 2008, 150, 247–254.
- Perera, E.; Moyano, F.J.; Díaz, M.; Perdomo-Morales, R.; Montero, V.; Rodríguez-Viera, L.; Alonso, E.; Carrillo, O.; Galich, G. Changes in digestive enzymes through developmental and molt stages in the spiny lobster, Panulirus argus. Comp. Biochem. Physiol. B 2008, 151, 250–256.
- Perera, E.; Rodríguez-Viera, L.; Perdomo-Morales, R.; Montero-Alejo, V.; Moyano, F.J.; Martínez-Rodríguez, G.; Mancera, J.M. Trypsin isozymes in the lobster Panulirus argus (Latreille, 1804): From molecules to physiology. J. Comp. Physiol. B 2015, 185, 17–35.
- Perera, E.; Rodríguez-Viera, L.; Rodríguez-Casariego, J.; Fraga, I.; Carrillo, O.; Martínez-Rodríguez, G.; Mancera, J.M. Dietary protein quality differentially regulates trypsin enzymes at the secretion and transcription level in Panulirus argus by distinct signaling pathways. J. Exper. Biol. 2012, 215, 853–862.
- Perera, E.; Simon, C. Digestive physiology of spiny lobsters: Implications for formulated diet development. Rev. Aquac. 2014, 6, 1–19.
- Rodríguez-Viera, L.; Perera, E.; Casuso, A.; Perdomo-Morales, R.; Gutierrez, O.; Scull, I.; Carrillo, O.; Martos-Sitcha, J.A.; García-Galano, T.; Mancera, J.M. A holistic view of dietary carbohydrate utilization in lobster: Digestion, postprandial nutrient flux, and metabolism. PLoS ONE 2014, 9, e108875.
- Rodríguez-Viera, L.; Perera, E.; Martos-Sitcha, J.A.; Perdomo-Morales, R.; Casuso, A.; Montero-Alejo, V.; García-Galano, T.; Martínez-Rodríguez, G.; Mancera, J.M. Molecular, biochemical, and dietary regulation features of α-Amylase in a carnivorous crustacean, the spiny lobster Panulirus argus. PLoS ONE 2016, 11, e0158919.
- Rodríguez-Viera, L.; Perera, E.; Montero-Alejo, V.; Perdomo-Morales, R.; García-Galano, T.; Martínez-Rodríguez, G.; Mancera, J.M. Carbohydrates digestion and metabolism in the spiny lobster (Panulirus argus): Biochemical indication for limited carbohydrate utilization. PeerJ 2017, 5, e3975.
- Johnston, D.J.; Freeman, J. Dietary preference and digestive enzyme activities as indicators of trophic resource utilization by six species of crab. Biol. Bull. 2005, 208, 36–46.
- Linton, S.M.; Greenaway, P. A review of feeding and nutrition of herbivorous land crabs: Adaptations to low quality plant diets. J. Comp. Physiol. B 2007, 177, 269–286.
- Linton, S.M. Presence and activity of endo-β-1,4-mannase, an important digestive carbohydrase within the digestive fluid of terrestrial crustaceans. J. Comp. Physiol. B 2021, 191, 243–253.
- Perera, E.; Fraga, I.; Carrillo, O.; Díaz-Iglesias, E.; Cruz, R.; Báez, M.; Galich, G. Evaluation of practical diets for the Caribbean spiny lobster Panulirus argus (Latreille, 1804): Effects of protein sources on substrate metabolism and digestive proteases. Aquaculture 2005, 244, 251–262.
- Perera, E.; Díaz-Iglesias, E.; Fraga, I.; Carrillo, O.; Galich, G. Effect of body weight, temperature and feeding on the metabolic rate in the spiny lobster Panulirus argus (Latreille, 1804). Aquaculture 2007, 265, 261–270.
- Wang, X.; Li, E.; Chen, L. A review of carbohydrate nutrition and metabolism in crustaceans. N. Am. J. Aquac. 2016, 78, 178–187.
- Cruz-Suarez, L.E.; Ricque-Marie, D.; Pinal-Mansilla, J.D.; Wesche-Ebelling, P. Effect of different carbohydrate sources on the growth of P. vannamei: Economical impact. Aquaculture 1994, 123, 349–360.
- Cuzon, G.; Lawrence, A.; Gaxiola, G.; Rosas, C.; Guillaume, J. Nutrition of Litopenaeus vannamei reared in tanks or in ponds. Aquaculture 2004, 235, 513–551.
- Johnston, D. Ontogenetic changes in digestive enzyme activity of the spiny lobster, Jasus edwardsii (Decapoda; Palinuridae). Mar. Biol. 2003, 143, 1071–1082.
- Le Moullac, G.; Klein, B.; Sellos, D.; Van Wormhoudt, A. Adaptation of trypsin, chymotrypsin and a-amylase to casein level and protein source in the shrimp P. vannamei. J. Exp. Mar. Biol. Ecol. 1996, 208, 107–125.
- Wei, J.; Zhang, X.; Yu, Y.; Huang, H.; Li, F.; Xiang, J. Comparative transcriptomic characterization of the early development in Pacific white shrimp Litopenaeus vannamei. PLoS ONE 2014, 9, e106201.
- Christensen, B.; Lomholt, B. Amylase heterogeneity in Palaemonetes varians (leach) (Crustacea, Decapoda). Ophelia 1972, 10, 63–65.
- Da Lage, J.L.; Van Wormhoudt, A.; Cariou, M.L. Diversity and evolution of the alpha-amylase genes in animals. Biol. Bratisl. 2002, 57, 181–190.
- Bloor, M.C. Dietary preference of Gammarus pulex and Asellus aquaticus during a laboratory breeding programme for ecotoxicological studies. Inter. J. Zool. 2011, 2011, 294394.
- Janeček, Š. How many conserved sequence regions are there in the a-amylase family? Biologia 2002, 57, 29–41.
- Grossman, G.L.; James, A.A. The salivary gland of the vector mosquito, Aedes aegypti, express a novel member of the amylase gene family. Insect. Mol. Biol. 1993, 1, 223–232.
- Froystad, M.K.; Lilleeng, E.; Sundby, A.; Krogdahl, A. Cloning and characterization of α-amylase from Atlantic salmon (Salmo salar L.). Comp. Biochem. Physiol. A 2006, 145, 479–492.
- Chávez-Rodríguez, L.; Rodríguez-Viera, L.; Montero-Alejo, V.; Perdomo-Morales, R.; Mancera, J.M.; Perera, E. A very active α-amylase and an inhibitor-based control of proteinases are key features of digestive biochemistry of the omnivorous Caribbean King Crab Maguimithrax spinosissimus. J. Evol. Biochem. Physiol. 2020, 56, 550–564.
- Vega-Villasante, F.; Nolasco, H.; Civera, R. The digestive enzymes of the Pacific brown shrimp Penaeus californiensis: I—Properties of amylase activity in the digestive tract. Comp. Biochem. Physiol. B 1993, 106, 547–550.
- Wojtowicz, M.B.; Brockerhoff, H. Isolation and some properties of the digestive amylase of the American lobster (Homarus americanus). Comp. Biochem. Physiol. B 1972, 42, 295–302.
- Guarna, M.M.; Borowsky, R.L. Biochemical properties of amylases from Gammarus palustris. Comp. Biochem. Physiol. B 1995, 112, 619–628.
- Biesiot, P.M.; Capuzzo, J.M. Changes in digestive enzyme activities during early development of the American lobster Homarus americanus Milne Edwards. J. Exp. Mar. Biol. Ecol. 1990, 136, 107–122.
- Janecek, Š. α-Amylase family: Molecular biology and evolution. Prog. Biophys. Mol. Biol. 1997, 67, 67–97.
- Aghajari, N.; Feller, G.; Gerday, C.; Haser, R. Structural basis of α-amylase activation by chloride. Protein Sci. 2002, 11, 1435–1441.
- Blandamer, A.; Beechey, R.B. The purification and properties of an alpha-amylase from the hepatopancreas of Carcinus maenas, the common shore crab. Biochim. Biophys. Acta 1966, 118, 204–206.
- Figueiredo, M.S.R.B.; Kricker, J.A.; Anderson, A.J. Digestive enzyme activities in the alimentary tract of redclaw crayfish, Cherax quadricarinatus (Decapoda: Parastacidae). J. Crust. Biol. 2001, 21, 334–344.
- Louati, H.; Zouari, N.; Fendri, A.; Gargouri, Y. Digestive amylase of a primitive animal, the scorpion: Purification and biochemical characterization. J. Chromatogr. B 2010, 878, 853–860.
- Žóltowska, K. The isoenzymes of o-amylase from the intestine of Ascaris suum. Helminthologia 2001, 38, 205–209.
- Lombraña, M.; Suárez, P.; San Juan, F. Two forms of α-amylase in mantle tissue of Mytilus galloprovincialis: Purification and molecular properties of form II. Comp. Biochem. Physiol. B 2005, 142, 56–66.
- Maalej, H.; Maalej, A.; Affes, S.; Hmidet, N.; Nasri, M.A. Novel Digestive α-Amylase from Blue Crab (Portunus segnis) Viscera: Purification, biochemical characterization and application for the improvement of antioxidant potential of oat flour. Int. J. Mol. Sci. 2021, 22, 1070.
- Dutta, T.K.; Jana, M.; Pahari, P.R.; Bhattacharya, T. The effect of temperature, pH, and salt on amylase in Heliodiaptomus viduus (Gurney) (Crustacea: Copepoda: Calanoida). Turk. J. Zool. 2006, 30, 187–195.
- Pavasovic, M.; Richardson, N.A.; Anderson, A.J.; Mann, D.; Mather, P.B. Effect of pH, temperature and diet on digestive enzyme profiles in the mud crab, Scylla serrata. Aquaculture 2004, 242, 641–654.
- Van Wormhoudt, A.; Bourreau, G.; Le Moullac, G. Amylase polymorphism in Crustacea Decapoda electrophoretic and immunological studies. Biochem. Syst. Ecol. 1995, 23, 139–149.
- Jones, D.A.; Kumlu, M.; Le Vay, L.; Fletcher, D.J. The digestive physiology of herbivorous, omnivorous and carnivorous crustacean larvae: A review. Aquaculture 1997, 155, 285–295.
- Kamarudin, M.S.; Jones, D.A.; Le Vay, L.; Abidin, Z. Ontogenetic change in digestive enzyme activity during larval development of Macrobrachium rosenbergii. Aquaculture 1994, 123, 323–333.
- Serrano Jr, A.E.; Traifalgar, R.F. Ontogeny and induction of digestive enzymes in Scylla serrata larvae fed live or artificial feeds or their combination. AACL Bioflux. 2012, 5, 101–111.
- Chen, J.; Chen, C.; Tan, O. Ontogenic changes in the digestive enzyme activities and the effect of different starvation duration on the digestive enzyme activities of larval red swamp crayfish (Procambarus clarkii). Aquac. Res. 2018, 49, 676–683.
- Le Vay, L.; Jones, D.A.; Puello-Cruz, A.C.; Sangha, R.S.; Ngamphongsai, C. Digestion in relation to feeding strategies exhibited by crustacean larvae. Comp. Biochem. Physiol. A 2001, 128, 623–630.
- Lovett, D.L.; Felder, D.L. Ontogenetic change in digestive enzyme activity of larval and postlarval white shrimp Penaeus setiferus (Crustacea, Decapoda, Penaeidae). Biol. Bull. 1990, 178, 144–159.
- Ribeiro, F.; Jones, D.A. Growth and ontogenetic change in activities of digestive enzymes in Fennero Penaeus indicus postlarvae. Aquac. Nutr. 2000, 6, 53–64.
- Musin, G.E.; Rossi, A.; Diawol, V.P.; Collins, P.A.; Williner, V. Development of enzymes during ontogeny of two freshwater Decapoda: Aegla uruguayana (Aeglidae) and Macrobrachium borellii (Palaemonidae). Aquac. Res. 2018, 49, 3889–3897.
- Figueiredo, M.S.R.B.; Anderson, A.J. Ontogenetic changes in digestive proteases and carbohydrases from the Australian freshwater crayfish, redclaw Cherax quadricarinatus (Crustacea, Decapoda, Parastacidae). Aquac. Res. 2003, 34, 1235–1239.
- Bergerson, O.; Wool, D. The process of adaptation of flour beetles to new environments. Genetica 1988, 77, 3–13.
- Inomata, N.; Nakashima, S. Short 5′-flanking regions of the Amygene of Drosophila kikkawai affect amylase gene expression and respond to food environments. Gene 2008, 412, 102–109.
- Prudence, M.; Moal, J.; Boudry, P.; Daniel, J.Y.; Quere, C.; Jeffroy, F.; Mingant, C.; Ropert, M.; Bédier, E.; Van Wormhoudt, A.; et al. An amylase gene polymorphism is associated with growth differences in the Pacific cupped oyster Crassostrea gigas. Anim. Genet. 2006, 37, 348–351.
- Hidalgo, M.C.; Urea, E.; Sanz, A. Comparative study of digestive enzymes in fish with different nutritional habits. Proteolytic and amylase activities. Aquaculture 1999, 170, 267–283.
- German, D.P.; Nagle, B.C.; Villeda, J.M.; Ruiz, A.M.; Thomson, A.W.; Contreras Balderas, S.; Evans, D.H. Evolution of herbivory in a carnivorous clade of minnows (Teleostei: Cyprinidae): Effects on gut size and digestive physiology. Physiol. Biochem. Zool. 2010, 83, 1–18.
- Axelsson, E.; Ratnakumar, A.; Arendt, M.L.; Maqbool, K.; Webster, M.T.; Perloski, M.; Liberg, O.; Arnemo, J.M.; Hedhammar, A.; Lindblad-Toh, K. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature 2013, 495, 360–364.
- Perry, G.H.; Dominy, N.J.; Claw, K.G.; Lee, A.S.; Fiegler, H.; Redon, R.; Werner, J.; Villanea, F.A.; Mountain, J.L.; Misra, R.; et al. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 2007, 39, 1256–1260.
- Karasov, W.H.; Douglas, A.E. Comparative digestive physiology. Compr. Physiol. 2013, 3, 741.
- Sather, B.T. A comparative study of amylases and proteinases in some decapod crustacea. Comp. Biochem. Physiol. 1969, 28, 371–379.
- Figueiredo, M.S.R.B.; Anderson, A.J. Digestive enzyme spectra in crustacean decapods (Paleomonidae, Portunidae and Penaeidae) feeding in the natural habitat. Aquac. Res. 2009, 40, 282–291.
- Huvet, A.; Jeffroy, F.; Fabioux, C.; Daniel, J.Y.; Quillien, V.; Van Wormhoudt, A.; Moal, J.; Samain, J.F.; Boudry, P.; Pouvreau, S. Association among growth, food consumption-related traits and amylase gene polymorphism in the Pacific oyster Crassostrea gigas. Anim. Genet. 2008, 39, 662–665.
- Omondi, J.G.; Stark, J.R. Some digestive carbohydrases from the midgut gland of Penaeus indicus and Penaeus vannamei (Decapoda: Penaeidae). Aquaculture 1995, 134, 121–135.
- Linton, S.M.; Saborowski, R.; Shirley, A.J.; Penny, J.A. Digestive enzymes of two brachyuran and two anomuran land crabs from Christmas Island, Indian Ocean. J. Comp. Physiol. B 2014, 184, 449–468.
- Simon, C.J. The effect of carbohydrate source, inclusion level of gelatinised starch, feed binder and fishmeal particle size on the apparent digestibility of formulated diets for spiny lobster juveniles, Jasus edwardsii. Aquaculture 2009, 296, 329–336.
- Wang, S.; Carter, C.G.; Fitzgibbon, Q.P.; Codabaccus, B.M.; Smith, G.G. Effect of dietary protein on energy metabolism including protein synthesis in the spiny lobster Sagmariasus verreauxi. Sci. Rep. 2021, 3, 11–11814.
- Hirche, H.J.; Anger, K. Digestive enzyme activities during larval development of Hyasaraneus (Decapoda, Majidae). Comp. Biochem. Physiol. B 1987, 87, 297–302.
- Chan, A.S.; Horn, M.H.; Dickson, K.A.; Gawlicka, A. Digestive enzyme activities in carnivores and herbivores: Comparisons among four closely related prickle back fishes (Teleostei: Stichaeidae) from a California rocky intertidal habitat. J. Fish. Biol. 2004, 65, 848–858.




