
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Agnieszka Barbara Najda | + 2951 word(s) | 2951 | 2021-09-28 04:33:25 | | | |
| 2 | Jason Zhu | Meta information modification | 2951 | 2021-10-07 05:49:02 | | | | |
| 3 | Agnieszka Barbara Najda | -291 word(s) | 2660 | 2021-10-07 12:53:01 | | | | |
| 4 | Conner Chen | Meta information modification | 2660 | 2021-10-11 08:58:48 | | | | |
| 5 | Jason Zhu | Meta information modification | 2660 | 2021-10-27 11:50:50 | | |
Video Upload Options
Adductomics is a transformative biomedical research tool that uses an "omic" approach to characterize and quantify exogenous and endogenous reactive compounds to which an individual is exposed; the use of compound-specific adduct biomarkers. Exposure to chemicals is generally driven by a variety of factors such as environment, genetics and lifestyle, which are characterized by a high level of interpersonal variability and contain a life element that makes it unique to each individual.
1. Wstęp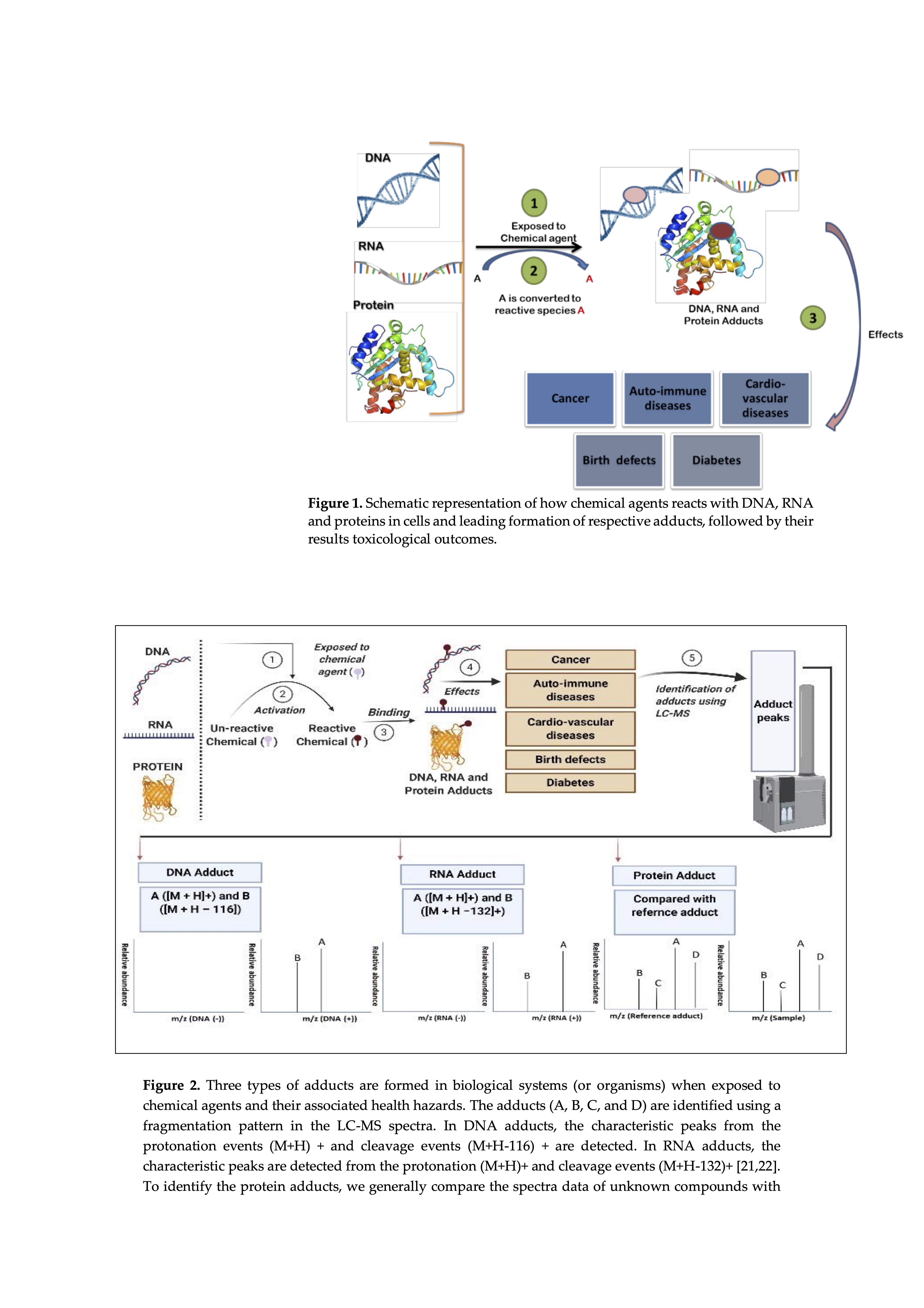
Adductomics has become the most significant technique in recent times and is one of the rapidly developing disciplines that has the potential to radically change the toxicology research landscape. It's probably time for adductomics to join the elite club of words like genomics, proteomics, and metabolomics. Although the term adductomics first appeared in a journal published in 2006 [ 1 ], it is now used in almost all areas of toxicological research. Adductomics is a transformative biomedical research tool that uses an "omic" approach to characterize and quantify exogenous and endogenous reactive compounds to which an individual is exposed;the use of compound-specific adduct biomarkers. Exposure to chemicals is generally driven by a variety of factors such as environment, genetics and lifestyle, which are characterized by a high level of interpersonal variability and contain a life element that makes it unique to each individual.
Adductomics focuses mainly on the study of adducts resulting from covalent modifications that are naturally irreversible with bio-macromolecules by exogenous or endogenous reactive electrophilic compounds. The reactive compounds interact with nucleophilic hot spots (electrophile susceptible sites) present in DNA, lipids, proteins, RNA and other macromolecules, leading to adduct formation (as shown in Figure 1 ). Biomonitoring of reactive metabolites regardless of their origin, whether exogenous or endogenous, is challenging due to their short in vivo lifetime, and adductomics has provided a unique opportunity to detect covalent adducts that are relatively stable and long-lived.Adductomy research makes extensive use of two approaches (1) targeted and (2) non-targeted; The first method focuses on the detection of specific adducts after exposure to specific chemical agents, and the later category aims to characterize all adducts by covalent bonds [ 2 , 3 ].
In some cases, natural chemicals do not bind directly to biological molecules, forming adducts, however, transformation into reactive metabolites occurs through metabolic enzymes such as cytochrome P450 systems, and the reactive metabolites formed may bind to protein, RNA and DNA [ 4 ]. , 5 ]. Reactive electrophiles generated from covalent genotoxic bonds with DNA can occur through several mechanisms: (1) arylamation [ 6 ], (2) alkylation [ 7 ], (3) development of bis-electrophilic cross-linking [ 8 ] and (4) adducts with highly reactive intermediates produced by lipid peroxidation [ 9 , 10, 11 ] or reactive oxygen species. The type and nature of the DNA adducts formed is usually dependent on many factors such as the chemical structure of the reactive chemicals, the ability of the chemical to intercalate with DNA, and the nature of the electrophiles. Evidence for aflatoxin, tobacco specific nitrosamine (NNK), polycyclic aromatic hydrocarbons (B [a] P), heterocyclic aromatic amine and other exogenous toxins create a different type of DNA adduct due to their different structural and chemical properties [ 12 ]. The formation of adducts may have a significant impact on the biological system and lead to harmful health complications [ 13] such as diabetes, neurodegenerative diseases, autoimmune diseases, cancer, birth defects [ 14 ] and cardiovascular diseases [ 15 ]. Understanding chemically induced adducts with adductomics may be essential to gain better insight into these diseases and provide new information on effective drug development. Adductomics can have a variety of practical applications in various fields, and its role is obvious (1) disease prognosis, (2) environmental health assessment [ 14 ], (3) development of personalized and precise medicine [ 16 ], (4) biomarker detection for various chemical exposures [ 17 , 18 ,19 ]. Measurement of DNA adducts resulting from exposure to a potential carcinogen in target organs is one of the basic methods of assessing the genotoxic potential of a chemical and serves as the most advanced method for determining the genotoxicity potential of a chemical. In addition, adductomics also identifies the underlying risk factors for pathogenesis and the underlying molecular mechanisms of chemically induced toxicity. The adductomics data would also serve as a guide for regulatory agencies and enable other stakeholders to take preventive measures against exposure to a toxic chemical.
Rapid refinement of methods and tools for the identification and quantification of adducts has made adductomics one of the most promising toxicological disciplines. Tissue sampling and sample preparation is one of the key factors in the detection of various adducts. Recent scientific advances and increased precision of the methods of detection and preparation of samples allow for their non-invasive sampling and the use of body fluids (blood plasma or serum, urine). liquid biopsy. The use of non-invasive methods such as liquid sampling provides various advantages such as 1) samples can be taken at different time intervals without causing great discomfort to patients 2) easy to collect and store 3) easy to transport. twenty]. Among the tools, a widely used and appropriate method for the qualitative and quantitative evaluation of adduct formation, including the identification of covalent conjugate sites in bionucleophiles, is high resolution mass spectrometry (HRMS). The fragmentation pattern in mass spectrometry is used to identify different types of adducts, and a notable aspect regarding DNA and RNA adducts is the near-universal loss of ribose and deoxyribose from the parent molecule giving characteristic peaks at (M + H-116) + and (M + H− 132) + respectively [ 20 , 21 ]. On the other hand, unknown protein adducts are identified by comparing the spectral data of the tested adducts with the reference adduct ( Figure 2 ).First, reference adducts should be synthesized assuming a specific electrophile, and then they will be matched with new adducts being the subject of further research. By adding the proposed electrophilic precursors to plasma or whole blood / lysate, reference adducts can be generated and fragmented using LC-MS. The synthetic adducts will then be further compared to new or unidentified adducts with m / z precursor ions, also examining fragmentation patterns and retention times. Moreover, this approach also contributes to the generation of an extensive database of reference protein adducts, thanks to which the identification of unknown protein adducts becomes much easier [ 22 ].
2. Application of adductomics
Adductomics is used in the assessment of pollutants and provides information on their toxic effects on biological systems that signal environmental health. For example, polycyclic aromatic hydrocarbons (PAHs), a toxic pollutant, are a group of structurally similar hydrocarbons released into the atmosphere as a result of incomplete combustion of organic matter, tobacco smoke, urban air pollution and car exhaust emissions [ 29 ]. PAHs can adducts with DNA via reactive intermediates when they are activated with cytochrome P-450 systems, which makes them highly carcinogenic [ 30]. ].One such reactive electrophilic form formed by CYP 1A1 and CYP 1B1 is the PAH-dihydrodiol epoxide, which can react with exocyclic groups present in nucleotides, such as guanine, adenine and cytosine present in DNA [ 31 ]. Similarly, many PAH-DNA adducts are formed with other reactive intermediates in individuals exposed to PAH, and the formed DNA adducts are examined by 32P-Post and LC-MS labeling [ 30 ]. Simultaneous assessment of the entire pool of PAH-DNA adducts in individuals provides us with a comprehensive exposure profile and facilitates a better understanding of the basic mechanistic pathways [ 32]. ].Another study established the relationship between the formation of PAH-DNA adducts in air pollution in exposed mothers and newborns in Poland, which can be seen in the dose-response curve, which showed a proportional increase in the number of DNA adducts with the degree of air pollution [ 33 ]. In the Mediterranean population, high-volume adducts are correlated with environmental pollution by ozone, which contributes to the formation of photochemical smog [ 34 ]. Hylland et al. [ 35 ] used DNA adducts as a distinctive biomarker to study the degree of contamination at various locations in the North East Atlantic region near the coast and at sea.The adduct as a biomarker (DNA adduct) alerts exposure to risk by providing early warning information and helping to improve aquatic hazard assessment and ecological risk assessment [ 36] ]. It was also revealed that DNA adducts (PAH-DNA adducts) would also help determine the biologically effective dose of PAH exposure, informing about the presence and extent of environmental contamination and its relationship to cancer development. PAHs are ubiquitous and their presence in oil and gas mixtures contaminates the aquatic ecosystem during oil and gas exploration. The detection of PAH-DNA adducts may also serve as potential biomarkers of environmental contamination and genotoxicity studies in aquatic organisms [ 37 ].In addition, several reports have shown evidence of the effects of crude oil and production gas on DNA adduct formation in marine organisms both in laboratory animals and in vivo following large oil spills [ 38 , 39 , 40 ].
Moreover, the detection of oxaplatin-induced DNA adducts in colorectal cancer patients with FOLFOX (a combination drug therapy containing folinic acid, fluorouracil and oxaliplatin) will help to design and optimize better treatment strategies for cancer patients. After FOLFAX treatment, the detected oxaplatin-DNA adducts in PBMCs were proportional to tumor reduction, making Drug-DNA adducts a potential biomarker in cancer treatment [ 50 ].
DNA adducts are physical complexes formed with DNA as a result of the interaction of reactive chemical species with DNA, and the detection of these adducts would serve as potential markers to determine a "biologically effective dose" for the presence of carcinogens in tobacco smoke and could help to better monitor the health of smokers. Several studies have shown that exposure to tobacco smoke can potentially induce DNA adduct formation in in vivo studies and have shown a positive correlation with carcinogenesis. In addition, the detection of DNA adducts can also provide a comprehensive measurement of exposure to carcinogens, also in cancer risk assessment and prediction.Several clinical and epidemiological studies have established an association between increased levels of DNA adducts and the likelihood of developing tobacco-related cancers such as lung, head, neck and bladder cancer [ 52 , 53 ]. While the DNA adduct profile provides exposure images, DNA adduct loading assesses the risk of carcinogenesis. The induction of DNA adducts in blood lymphocytes is also believed to be associated with the development of head and neck cancer.However, the dose-response relationship between smoking and DNA adducts in exposed organs is not fully characterized and in fact the relationship is complicated due to inconsistencies in epidemiological studies and a genetic polymorphism (in carcinogen metabolism (e.g. GSTP1) and DNA repair (e.g. XRCC1)) is the root cause. In early-stage tumors of tobacco carcinogenesis induced by p53 mutations and DNA adducts, it was observed that levels of DNA adducts were correlated with somatic changes (eg 3p21 LOH) [ 69 ].
Rating genetic toxicity has a high priority in managing safety risks during the development of new chemical compounds and it does so by assessing the carcinogenicity and mutagenicity of the chemical, thus helping in the hazard identification and risk characteristics of chemical agents [ 97 , 98 ]. Traditionally potential genotoxicity, carcinogenicity, and mutagenicity of chemical is evaluated by the Ames test, a test, micronucleus test and the chromosomal [ 99 , 100 ].However, the challenge with these in vitro methods is the high false-positive rate that requires the development of new methodologies and pathway-based understanding of toxicity, which can provide a more accurate picture of DNA damage that can directly detect DNA modifications and DNA damage at the molecular level [ 101 , 102 , 103 ]. Here, DNA adductomics turns out to be a potential candidate for a methodology that could comprehensively study DNA damage through direct molecular detection through the identification and quantification of DNA adducts [ 104 , 105 , 106 ].The micronucleus test is one of the widely used in vitro tests to assess DNA damage, but is currently supplemented by DNA adductomy to address the error due to false-positive test results, confirming the role of DNA adductomics in other in vitro genotoxicity assessment [ 107 , 108 ].
3. Other Applications
In addition to the above, other uses of adductomics are summarized in Table 1 below along with publication titles along with novel uses of adductomics:
4. Current challenges and prospects for the future
Advances in diagnostic tools and the emergence of new technologies have given rise to the use of adductomics. However, there are still challenges that need to be overcome in order to fully exploit the potential of adductomics in the toxicological and environmental assessment of chemicals. While data-driven and data-independent extraction methods (in untargeted "omics") have been developed to test multiple adducts simultaneously, obstacles to data processing need to be addressed to get an accurate picture of toxic substances [ 104]. ]. The low frequency of DNA adducts in the sample pool also poses a serious challenge to current software in a realistic assessment that uses common data acquisition methods.This is the need for the continuation of data processing software and the improvement of algorithms for detecting adducts, even at low concentrations, which are critical to understanding pathogenesis [ 143] ]. There are a number of improvements in sample preparation and purification regarding the detection of hydrophilic adducts. Moreover, incomplete enzymatic hydrolysis does not generate and observe certain types of DNA adducts, requiring a comprehensive evaluation of the advantages and disadvantages of several enzymes in terms of DNA hydrolysis and their optimal use.In adducts with molecular weights below 70 KDa there are few probable structures and their identification is not troublesome, while in adducts with higher molecular weights their characterization is extremely difficult due to extended possibilities and amplified permutations; this is a matter of concern even if we could make accurate mass measurements and generate ion fragmentation spectra. This handicap can be overcome by creating an adduct database that would provide ready-made information about adducts;Unfortunately, there is no specific database for adductomics, although hundreds of DNA adducts are characterized every day around the world, the creation of such a database requires a thorough literature search of molecular formulas of already characterized adducts. Fragmentation spectra generated from both ion trap and quadrupole-type fragmentation at the MS2 and MS3 levels, demonstrated at different collision energies, would be useful if compiled and integrated into a database. Currently, databases such as Search for Species Data by Molecular Weight provided by NIST (National Institute of Standards and Technologies) [there is no specific database for adductomics, although hundreds of DNA adducts are characterized every day all over the world, the creation of such a database requires a thorough literature search of molecular formulas of already characterized adducts. Fragmentation spectra generated from both ion trap and quadrupole-type fragmentation at the MS2 and MS3 levels, demonstrated at different collision energies, would be useful if compiled and integrated into a database. Currently, databases such as Search for Species Data by Molecular Weight provided by NIST (National Institute of Standards and Technologies) [there is no specific database for adductomics, although hundreds of DNA adducts are characterized every day all over the world, the creation of such a database requires a thorough literature search of molecular formulas of already characterized adducts. Fragmentation spectra generated from both ion trap and quadrupole-type fragmentation at the MS2 and MS3 levels, demonstrated at different collision energies, would be useful if compiled and integrated into a database. Currently, databases such as Search for Species Data by Molecular Weight provided by NIST (National Institute of Standards and Technologies) [Fragmentation spectra generated from both ion trap and quadrupole-type fragmentation at the MS2 and MS3 levels, demonstrated at different collision energies, would be useful if compiled and integrated into a database. Currently, databases such as Search for Species Data by Molecular Weight provided by NIST (National Institute of Standards and Technologies) [ Fragmentation spectra generated from both ion trap and quadrupole-type fragmentation at MS2 and MS3 levels, shown at different collision energies, would be useful if compiled and integrated with the database. Currently, databases such as Search for Species Data by Molecular Weight provided by NIST (National Institute of Standards and Technologies) [ 146], UNIMOD [ 147 ], Human Metabolome Database [ 142 ], Toxic Exposure Database [ 143 ], Exposome- Explorer Database [ 144] ] find use in adductomics. However, the above-mentioned databases are not adductomics specific, which requires the creation of a dedicated database that can facilitate easy identification of unknown adducts. There is a need to develop a more robust and simple technology to further refine sampling, as suggested above, a more focused approach to non-invasive liquid sampling, optimization of sample preparation methods is required that can give precise and reproducible results.Current analytical techniques are very time consuming and expensive to test samples, and further development of cost effective analytical techniques could further enhance the applications of adductomics in biomedical research.
The entry is from 10.3390/ijms221810141
References
- Kanaly, R.A.; Hanaoka, T.; Sugimura, H.; Toda, H.; Matsui, S.; Matsuda, T. Development of the Adductome Approach to Detect DNA Damage in Humans. Antioxidants Redox Signal. 2006, 8, 993–1001.
- Carlsson, H.; Rappaport, S.M.; Törnqvist, M. Protein Adductomics: Methodologies for Untargeted Screening of Adducts to Serum Albumin and Hemoglobin in Human Blood Samples. High-Throughput 2019, 8, 6.
- Preston, G.W.; Phillips, D.H. Protein Adductomics: Analytical Developments and Applications in Human Biomonitoring. Toxics 2019, 7, 29.
- Guengerich, F.P. Common and Uncommon Cytochrome P450 Reactions Related to Metabolism and Chemical Toxicity. Chem. Res. Toxicol. 2001, 14, 611–650.
- Rendic, S.; Guengerich, F.P. Contributions of Human Enzymes in Carcinogen Metabolism. Chem. Res. Toxicol. 2012, 25, 1316–1383.
- Beland, F.A.; Beranek, D.T.; Dooley, K.L.; Heflich, R.H.; Kadlubar, F.F. Arylamine-DNA adducts in vitro and in vivo: Their role in bacterial mutagenesis and urinary bladder carcinogenesis. Environ. Health Perspect. 1983, 49, 125–134.
- Shrivastav, N.; Li, D.; Essigmann, J.M. Chemical biology of mutagenesis and DNA repair: Cellular responses to DNA alkylation. Carcinogenesis 2009, 31, 59–70.
- Rajski, S.R.; Williams, R.M. DNA Cross-Linking Agents as Antitumor Drugs. Chem. Rev. 1998, 98, 2723–2796.
- Marnett, L.J. Lipid peroxidation—DNA damage by malondialdehyde. Mutat. Res. Mol. Mech. Mutagen. 1999, 424, 83–95.
- Tudek, B.; Zdżalik-Bielecka, D.; Tudek, A.; Kosicki, K.; Fabisiewicz, A.; Speina, E. Lipid peroxidation in face of DNA damage, DNA repair and other cellular processes. Free. Radic. Biol. Med. 2017, 107, 77–89.
- Marnett, L.J. Oxy radicals, lipid peroxidation and DNA damage. Toxicology 2002, 181, 219–222.
- Guo, J.; Turesky, R.J. Emerging Technologies in Mass Spectrometry-Based DNA Adductomics. High-Throughput 2019, 8, 13.
- Colombo, G.; Clerici, M.; Giustarini, D.; Rossi, R.; Milzani, A.D.G.; Dalle-Donne, I. Redox Albuminomics: Oxidized Albumin in Human Diseases. Antioxid. Redox Signal. 2012, 17, 1515–1527.
- Gorokhova, E.; Martella, G.; Motwani, N.H.; Tretyakova, N.Y.; Sundelin, B.; Motwani, H.V. DNA epigenetic marks are linked to embryo aberrations in amphipods. Sci. Rep. 2020, 10, 1–11.
- Zhang, H.; Ge, Y. Comprehensive Analysis of Protein Modifications by Top-Down Mass Spectrometry. Circ. Cardiovasc. Genet. 2011, 4, 711.
- Stornetta, A.; Zimmermann, M.; Cimino, G.D.; Henderson, P.T.; Sturla, S.J. DNA Adducts from Anticancer Drugs as Candidate Predictive Markers for Precision Medicine. Chem. Res. Toxicol. 2017, 30, 388–409.
- Harris, C.C. Future directions in the use of DNA adducts as internal dosimeters for monitoring human exposure to environmental mutagens and carcinogens. Environ. Heal. Perspect. 1985, 62, 185–191.
- La, D.K.; Swenberg, J.A. DNA adducts: Biological markers of exposure and potential applications to risk assessment. Mutat. Res. Genet. Toxicol. 1996, 365, 129–146.
- Jarabek, A.M.; Pottenger, L.H.; Andrews, L.S.; Casciano, D.; Embry, M.R.; Kim, J.H.; Preston, R.J.; Reddy, M.V.; Schoeny, R.; Shuker, D.; et al. Creating context for the use of DNA adduct data in cancer risk assessment: I. Data organization. Crit. Rev. Toxicol. 2009, 39, 659–678.
- Balbo, S.; Turesky, R.J.; Villalta, P.W. DNA Adductomics. Chem. Res. Toxicol. 2014, 27, 356–366.
- Takeshita, T.; Kanaly, R.A. In vitro DNA/RNA Adductomics to Confirm DNA Damage Caused by Benzopyrene in the Hep G2 Cell Line. Front. Chem. 2019, 7, 7.
- Rappaport, S.M. Genetic Factors Are Not the Major Causes of Chronic Diseases. PLOS ONE 2016, 11, e0154387.
- Grimmer, V.G. Environmental Carcinogens: Polycyclic Aromatic Hydrocarbons; CRC Press: Boca Raton, FL, USA, 1983.
- Singh, R.; Teichert, F.; Seidel, A.; Roach, J.; Cordell, R.; Cheng, M.-K.; Frank, H.; Steward, W.P.; Manson, M.M.; Farmer, P.B. Development of a targeted adductomic method for the determination of polycyclic aromatic hydrocarbon DNA adducts using online column-switching liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2010, 24, 2329–2340.
- Wang, J.J.; Marshall, W.D.; Frazer, D.G.; Law, B.; Lewis, D.M. Characterization of DNA adducts from lung tissue of asphalt fume-exposed mice by nanoflow liquid chromatography quadrupole time-of-flight mass spectrometry. Anal. Biochem. 2003, 322, 79–88.
- Ewa, B.; Danuta, M.-. Švach Polycyclic aromatic hydrocarbons and PAH-related DNA adducts. J. Appl. Genet. 2017, 58, 321–330.
- Whyatt, R.M.; Santella, R.M.; Jedrychowski, W.; Garte, S.J.; Bell, D.; Ottman, R.; Gladek-Yarborough, A.; Cosma, G.; Young, T.L.; Cooper, T.B.; et al. Relationship between ambient air pollution and DNA damage in Polish mothers and newborns. Environ. Heal. Perspect. 1998, 106, 821–826.
- Palli, D.; Saieva, C.; Grechi, D.; Masala, G.; Zanna, I.; Barbaro, A.; Decarli, A.; Munnia, A.; Peluso, M. DNA bulky adducts in a Mediterranean population correlate with environmental ozone concentration, an indicator of photochemical smog. Int. J. Cancer 2004, 109, 17–23.
- Hylland, K.; Skei, B.B.; Brunborg, G.; Lang, T.; Gubbins, M.J.; le Goff, J.; Burgeot, T. DNA damage in dab (Limanda limanda) and haddock (Melanogrammus aeglefinus) from European seas. Mar. Environ. Res. 2017, 124, 54–60.
- A Hagger, J.; Jones, M.B.; Leonard, P.; Owen, R.; Galloway, T.S. Biomarkers and integrated environmental risk assessment: Are there more questions than answers? Integr. Environ. Assess. Manag. 2006, 2, 312–329.
- Pampanin, D.M.; Brooks, S.J.; Grøsvik, B.E.; Le Goff, J.; Meier, S.; Sydnes, M.O. DNA adducts in marine fish as biological marker of genotoxicity in environmental monitoring: The way forward. Mar. Environ. Res. 2017, 125, 49–62.
- Lyons, B.; Stewart, C.; Kirby, M. The detection of biomarkers of genotoxin exposure in the European flounder (Platichthys flesus) collected from the River Tyne Estuary. Mutat. Res. Toxicol. Environ. Mutagen. 1999, 446, 111–119.
- Aas, E.; Baussant, T.; Balk, L.; Liewenborg, B.; Andersen, O.K. PAH metabolites in bile, cytochrome P4501A and DNA adducts as environmental risk parameters for chronic oil exposure: A laboratory experiment with Atlantic cod. Aquat. Toxicol. 2000, 51, 241–258.
- Harvey, J.; Lyons, B.; Page, T.; Stewart, C.; Parry, J. An assessment of the genotoxic impact of the Sea Empress oil spill by the measurement of DNA adduct levels in selected invertebrate and vertebrate species. Mutat. Res. Toxicol. Environ. Mutagen. 1999, 441, 103–114.
- Zimmermann, M.; Li, T.; Semrad, T.J.; Wu, C.-Y.; Yu, A.; Cimino, G.; Malfatti, M.; Haack, K.; Turteltaub, K.W.; Pan, C.-X.; et al. Oxaliplatin–DNA Adducts as Predictive Biomarkers of FOLFOX Response in Colorectal Cancer: A Potential Treatment Optimization Strategy. Mol. Cancer Ther. 2020, 19, 1070–1079.
- Johnson, L.A.; Malayappan, B.; Tretyakova, N.; Campbell, C.; MacMillan, M.; Wagner, J.E.; Jacobson, P.A. Formation of cyclophosphamide specific DNA adducts in hematological diseases. Pediatr. Blood Cancer 2012, 58, 708–714.
- Stornetta, A.; Villalta, P.W.; Gossner, F.; Wilson, W.R.; Balbo, S.; Sturla, S.J. DNA Adduct Profiles Predict in Vitro Cell Viability after Treatment with the Experimental Anticancer Prodrug PR104A. Chem. Res. Toxicol. 2017, 30, 830–839.
- National Toxicology Program (NTP). Toxicology and Carcinogenesis Studies of Furan (CAS NO. 110-00-9) in F344/N Rats and B6C3F1 Mice (Gavage Studies); NTP TR 402 (NIH Publication No. 93-2857); U.S. Department of Health and Human Services, Public Health: Washington, DC, USA, 1993; pp. 1–287.
- Löf, M.; Sundelin, B.; Liewenborg, B.; Bandh, C.; Broeg, K.; Schatz, S.; Gorokhova, E. Biomarker-enhanced assessment of reproductive disorders in Monoporeia affinis exposed to contaminated sediment in the Baltic Sea. Ecol. Indic. 2016, 63, 187–195.
- Löf, M.F.; Sundelin, B.; Bandh, C.; Gorokhova, E. Embryo aberrations in the amphipod Monoporeia affinis as indicators of toxic pollutants in sediments: A field evaluation. Ecol. Indic. 2016, 60, 18–30.
- Cimino, M.C. Comparative overview of current international strategies and guidelines for genetic toxicology testing for regulatory purposes. Environ. Mol. Mutagen. 2006, 47, 362–390.
- Thybaud, V.; Lorge, E.; Levy, D.; van Benthem, J.; Douglas, G.R.; Marchetti, F.; Moore, M.M.; Schoeny, R. Main issues addressed in the 2014-2015 revisions to the OECD Genetic Toxicology Test Guidelines. Environ. Mol. Mutagen. 2017, 58, 284–295.
- Kirkland, D.; Aardema, M.; Henderson, L.; Müller, L. Evaluation of the ability of a battery of three in vitro genotoxicity tests to discriminate rodent carcinogens and non-carcinogens: I. Sensitivity, specificity and relative predictivity. Mutat. Res. Toxicol. Environ. Mutagen. 2005, 584, 1–256.
- Hayashi, M.; Honma, M.; Takahashi, M.; Horibe, A.; Tanaka, J.; Tsuchiya, M.; Morita, T. Identification and Evaluation of Potentially Genotoxic Agricultural and Food-related Chemicals. Food Saf. 2013, 1, 2013003.
- Kirkland, D.; Pfuhler, S.; Tweats, D.; Aardema, M.; Corvi, R.; Darroudi, F.; Elhajouji, A.; Glatt, H.; Hastwell, P.; Hayashi, M.; et al. How to reduce false positive results when undertaking in vitro genotoxicity testing and thus avoid unnecessary follow-up animal tests: Report of an ECVAM Workshop. Mutat. Res. Toxicol. Environ. Mutagen. 2007, 628, 31–55.
- Sobus, J.R.; Wambaugh, J.; Isaacs, K.; Williams, A.; McEachran, A.; Richard, A.M.; Grulke, C.; Ulrich, E.; Rager, J.E.; Strynar, M.; et al. Integrating tools for non-targeted analysis research and chemical safety evaluations at the US EPA. J. Expo. Sci. Environ. Epidemiology 2018, 28, 411–426.
- Kanaly, R.A.; Matsui, S.; Hanaoka, T.; Matsuda, T. Application of the adductome approach to assess intertissue DNA damage variations in human lung and esophagus. Mutat. Res. Mol. Mech. Mutagen. 2007, 625, 83–93.
- Kato, K.; Yamamura, E.; Kawanishi, M.; Yagi, T.; Matsuda, T.; Sugiyama, A.; Uno, Y. Application of the DNA adductome approach to assess the DNA-damaging capability of in vitro micronucleus test-positive compounds. Mutat. Res. Toxicol. Environ. Mutagen. 2011, 721, 21–26.
- Rappaport, S.M.; Smith, M.T. Environment and Disease Risks. Science 2010, 330, 460–461.
- HEP. The Human Exposome Project: A Toolbox for Assessing and Addressing the Impact of Environment on Health. Available online: https://cordis.europa.eu/programme/id/H2020_SC1-BHC-28-2019 (accessed on 10 July 2021).
- Toxic Exposome Database, T3DB. Available online: http://www.t3db.ca/ (accessed on 10 July 2021).
- Molecular Weight Search. Available online: http://webbook.nist.gov/chemistry/mw-ser.html (accessed on 10 July 2021).
- Unimod. Available online: http://www.unimod.org/modifications_list.php (accessed on 10 July 2021).
- Human Metabolome Database. Available online: https://hmdb.ca/ (accessed on 17 September 2021).
- Neveu, V.; Moussy, A.; Rouaix, H.; Wedekind, R.; Pon, A.; Knox, C.; Wishart, D.S.; Scalbert, A. Exposome-Explorer: A manually-curated database on biomarkers of exposure to dietary and environmental factors. Nucleic Acids Res. 2017, 45, D979–D984.




