
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Carlos Cabañas | + 3034 word(s) | 3034 | 2021-09-16 08:24:49 | | | |
| 2 | Jason Zhu | Meta information modification | 3034 | 2021-09-22 04:48:18 | | |
Video Upload Options
Exosomes are a type of extracellular vesicle (EV) of endocytic origin that are released by virtually all cells in multicellular organisms and carry out important intercellular communication functions through the transfer of their biomolecular cargo, which includes lipids, proteins, nucleic acids and metabolites, between the producing and the recipient/target cells [1]. Exosomes produced by cancer cells have been shown to influence many processes related to cancer progression and metastasis, such as tumor cell proliferation and invasion, angiogenesis, tumor microenvironment promotion and remodelling, chemotherapy resistance, and immune suppression (recently reviewed in [2,3,4]).
1. Introduction
Colorectal cancer (CRC) has a high incidence and is a major cause of cancer-related mortality worldwide, with approximately 25% of patients developing peritoneal metastasis, a condition associated with a bleak prognosis as current treatments are less effective in these patients [1][2][3][4][5]. The CRC peritoneal dissemination cascade involves sequential shedding of cancer cells from the primary colonic or rectal tumor, their transport through the peritoneal cavity following the physiological flow of peritoneal fluid, their adhesion to the peritoneal mesothelial cells (PMCs) that form a barrier lining all peritoneal organs, and the subsequent invasion of cancer cells through this mesothelial cell barrier and the underlying stroma to establish new metastatic foci in the peritoneum, omentum and bowel serosa. In addition to acting as a barrier, PMCs also play an active role in the pathogenesis of peritoneal metastasis by differentiating into cancer-associated fibroblasts (CAFs) through a process termed Mesothelial-to-Mesenchymal Transition (MMT), which fuels peritoneal metastasis and immune evasion (reviewed in [5]).
In epithelial ovarian cancer, which predominantly disseminates via peritoneal metastasis, exosomes produced by ovarian cancer cells favor different steps of the peritoneal dissemination cascade and have therefore been recognized to play crucial roles in the pathogenesis of the disease [6][7]. Of particular relevance, cancer-derived exosomes in malignant ascites interact with cancer cells promoting their survival, migration and invasion capacities, and with PMCs inducing their apoptosis, disrupting the mesothelial barrier and reprogramming them into CAFs through MMT. For CRC, however, current knowledge about the roles played by exosomes in the pathogenesis and peritoneal metastasis cascade is sparse. At the same time, the molecules that mediate the interactions between cancer-derived exosomes and their target PMCs and tumor cells mostly remain obscure.
Previously we employed the human colorectal adenocarcinoma Colo-320 cell line [8] as a useful model to study cancer-related processes such as tumorigenesis, metastasis, and tumor cell adhesion, as well as the functions and interplay between tetraspanins and cell adhesion molecules [9][10][11][12][13][14]. Colo-320 cells possess the distinctive feature of lacking endogenous expression of tetraspanin CD9 while they abundantly express integrin α5β1 but not many other members of the β1 integrin subfamily. CD9 is a widely distributed tetraspanin that associates with other transmembrane proteins, including integrins and ADAM metalloproteases in TEMs (Tetraspanin-Enriched Microdomains) (reviewed in [15]). CD9 is involved in cell adhesion, motility, sperm–egg fusion, tumorigenesis and metastasis and an inverse correlation between expression of CD9 and metastatic potential and patient survival rate has been established in many types of cancer, including CRC (reviewed in [16]).
Several reports indicate that integrin α5β1 can engage in cis (on the same cell) and trans (on different cells) interactions with the distintegrin (Dis) domain of the metalloproteinase ADAM17/TACE [15][17][18][19][20]. Recent study showed that the tetraspanin CD9 negatively regulated integrin α5β1-mediated adhesion of cancer cells both to its canonical ligand fibronectin and to ADAM17 as a novel ligand[14][15] Furthermore, it has reported that CD9 interacts directly with ADAM17 on the cell surface and regulates negatively both the sheddase and adhesive activities of this metalloproteinase [12][13][14][17][18][19][20][21]. Therefore, we decided to investigate whether integrin α5β1 and ADAM17 interaction and the regulatory effects of CD9 also played a role in mediating the binding of exosomes and their uptake by cancer or mesothelial recipient cells, which could bear relevance during the peritoneal dissemination of colorectal carcinomas.
We report here that the interaction between integrin α5β1 on colorectal carcinoma cells and on PMCs and its ligand ADAM17 on exosomes mediates the binding and uptake of cancer-derived exosomes. Furthermore, this process was negatively regulated by the expression of tetraspanin CD9 on the surface of exosomes.
2. Results
2.1. Characterization of EVs Derived from Colo-320, Colo-320/CD9 and Colo-320/ADAM17-KO Human Adenocarcinoma Cells
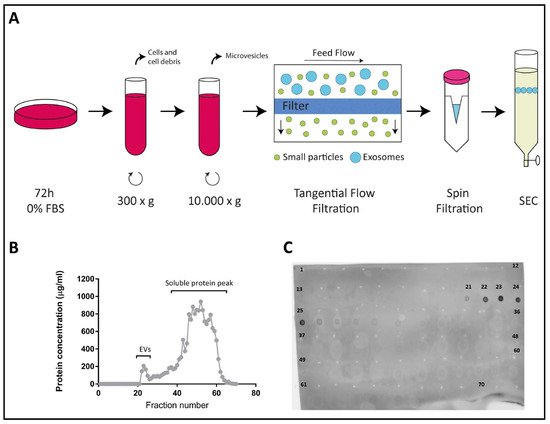
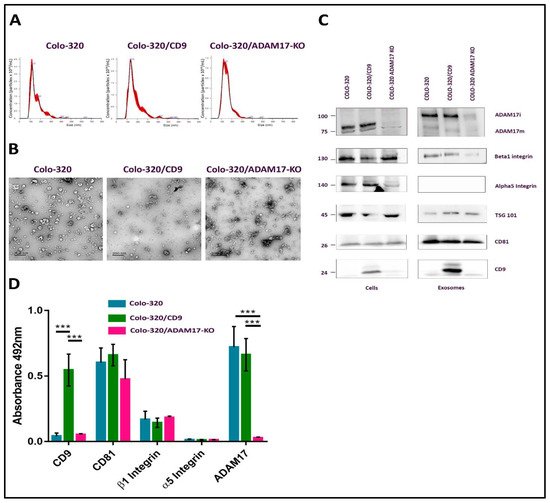
2.2. Fibronectin Is Not the Ligand of Integrin α5β1 That Mediates Interactions of Exosomes with Colo-320 Cancer Cells
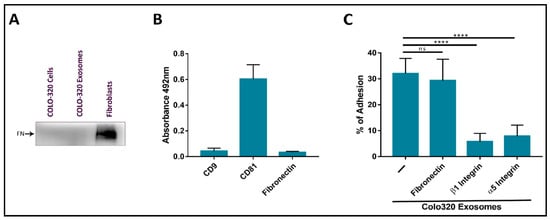
2.3. Expression of CD9 Reduces Interactions between Cancer Cells and Exosomes Mediated by Cellular Integrin α5β1 and Exosomal ADAM17
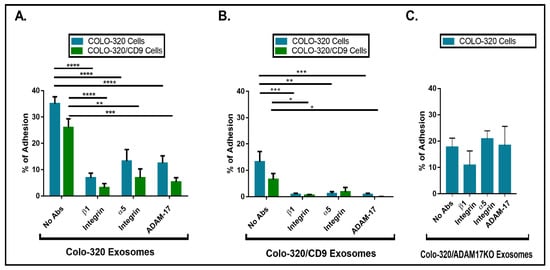
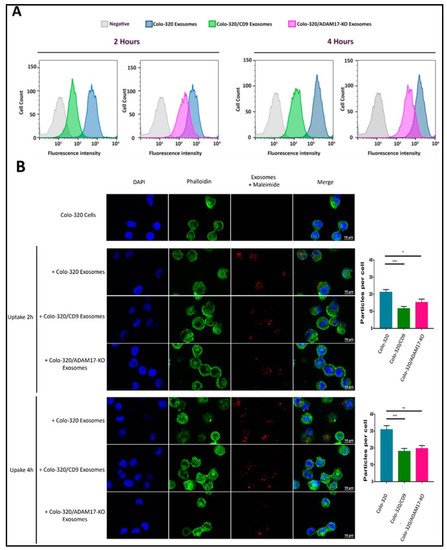
2.4. Uptake of Exosomes by Colo-320 Cancer Cells Depends on Exosomal ADAM17 and Is Inhibited by CD9
References
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, Y.; Ding, K. Roles of exosomes in cancer chemotherapy resistance, progression, metastasis and immunity, and their clinical applications (Review). Int. J. Oncol. 2021, 59, 1–18. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, K.; Chen, Y.; Wu, X.; Chen, Z.; Cao, K.; Tao, Y.; Chen, X.; Liao, J.; Zhou, J. Exosomes and Their Role in Cancer Progression. Front. Oncol. 2021, 11, 639159. [Google Scholar] [CrossRef] [PubMed]
- Willms, E.; Cabanas, C.; Mager, I.; Wood, M.J.A.; Vader, P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front. Immunol. 2018, 9, 738. [Google Scholar] [CrossRef] [PubMed]
- Ceelen, W.; Ramsay, R.G.; Narasimhan, V.; Heriot, A.G.; De Wever, O. Targeting the Tumor Microenvironment in Colorectal Peritoneal Metastases. Trends Cancer 2020, 6, 236–246. [Google Scholar] [CrossRef]
- Nakamura, K.; Sawada, K.; Kinose, Y.; Yoshimura, A.; Toda, A.; Nakatsuka, E.; Hashimoto, K.; Mabuchi, S.; Morishige, K.I.; Kurachi, H.; et al. Exosomes Promote Ovarian Cancer Cell Invasion through Transfer of CD44 to Peritoneal Mesothelial Cells. Mol. Cancer Res. MCR 2017, 15, 78–92. [Google Scholar] [CrossRef]
- Nakamura, K.; Sawada, K.; Kobayashi, M.; Miyamoto, M.; Shimizu, A.; Yamamoto, M.; Kinose, Y.; Kimura, T. Role of the Exosome in Ovarian Cancer Progression and Its Potential as a Therapeutic Target. Cancers 2019, 11, 1147. [Google Scholar] [CrossRef] [PubMed]
- Quinn, L.A.; Moore, G.E.; Morgan, R.T.; Woods, L.K. Cell lines from human colon carcinoma with unusual cell products, double minutes, and homogeneously staining regions. Cancer Res. 1979, 39, 4914–4924. [Google Scholar] [PubMed]
- Gutierrez-Lopez, M.D.; Ovalle, S.; Yanez-Mo, M.; Sanchez-Sanchez, N.; Rubinstein, E.; Olmo, N.; Lizarbe, M.A.; Sanchez-Madrid, F.; Cabanas, C. A functionally relevant conformational epitope on the CD9 tetraspanin depends on the association with activated beta1 integrin. J. Biol. Chem. 2003, 278, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Barreiro, O.; Yanez-Mo, M.; Sala-Valdes, M.; Gutierrez-Lopez, M.D.; Ovalle, S.; Higginbottom, A.; Monk, P.N.; Cabanas, C.; Sanchez-Madrid, F. Endothelial tetraspanin microdomains regulate leukocyte firm adhesion during extravasation. Blood 2005, 105, 2852–2861. [Google Scholar] [CrossRef] [PubMed]
- Ovalle, S.; Gutierrez-Lopez, M.D.; Olmo, N.; Turnay, J.; Lizarbe, M.A.; Majano, P.; Molina-Jimenez, F.; Lopez-Cabrera, M.; Yanez-Mo, M.; Sanchez-Madrid, F.; et al. The tetraspanin CD9 inhibits the proliferation and tumorigenicity of human colon carcinoma cells. Int. J. Cancer 2007, 121, 2140–2152. [Google Scholar] [CrossRef]
- Gutierrez-Lopez, M.D.; Gilsanz, A.; Yanez-Mo, M.; Ovalle, S.; Lafuente, E.M.; Dominguez, C.; Monk, P.N.; Gonzalez-Alvaro, I.; Sanchez-Madrid, F.; Cabanas, C. The sheddase activity of ADAM17/TACE is regulated by the tetraspanin CD9. Cell Mol. Life Sci. 2011, 68, 3275–3292. [Google Scholar] [CrossRef] [PubMed]
- Gilsanz, A.; Sanchez-Martin, L.; Gutierrez-Lopez, M.D.; Ovalle, S.; Machado-Pineda, Y.; Reyes, R.; Swart, G.W.; Figdor, C.G.; Lafuente, E.M.; Cabanas, C. ALCAM/CD166 adhesive function is regulated by the tetraspanin CD9. Cell Mol. Life Sci. 2013, 70, 475–493. [Google Scholar] [CrossRef] [PubMed]
- Machado-Pineda, Y.; Cardenes, B.; Reyes, R.; Lopez-Martin, S.; Toribio, V.; Sanchez-Organero, P.; Suarez, H.; Grotzinger, J.; Lorenzen, I.; Yanez-Mo, M.; et al. CD9 Controls Integrin alpha5beta1-Mediated Cell Adhesion by Modulating Its Association with the Metalloproteinase ADAM17. Front. Immunol. 2018, 9, 2474. [Google Scholar] [CrossRef]
- Reyes, R.; Cardenes, B.; Machado-Pineda, Y.; Cabanas, C. Tetraspanin CD9: A Key Regulator of Cell Adhesion in the Immune System. Front. Immunol. 2018, 9, 863. [Google Scholar] [CrossRef]
- Lorico, A.; Lorico-Rappa, M.; Karbanova, J.; Corbeil, D.; Pizzorno, G. CD9, a tetraspanin target for cancer therapy? Exp. Biol. Med. 2021, 246, 1121–1138. [Google Scholar] [CrossRef]
- Bax, D.V.; Messent, A.J.; Tart, J.; van Hoang, M.; Kott, J.; Maciewicz, R.A.; Humphries, M.J. Integrin alpha5beta1 and ADAM-17 interact in vitro and co-localize in migrating HeLa cells. J. Biol. Chem. 2004, 279, 22377–22386. [Google Scholar] [CrossRef]
- Grotzinger, J.; Lorenzen, I.; Dusterhoft, S. Molecular insights into the multilayered regulation of ADAM17: The role of the extracellular region. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 2088–2095. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, G.; Jia, N.; Wang, W.; Wang, Y.; Yin, M.; Jiang, X.; Huang, Y.; Zhang, J. CD9 regulates keratinocyte migration by negatively modulating the sheddase activity of ADAM17. Int. J. Biol. Sci. 2019, 15, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Mikulicic, S.; Fritzen, A.; Scheffer, K.; Strunk, J.; Cabanas, C.; Sperrhacke, M.; Reiss, K.; Florin, L. Tetraspanin CD9 affects HPV16 infection by modulating ADAM17 activity and the ERK signalling pathway. Med. Microbiol. Immunol. 2020, 209, 461–471. [Google Scholar] [CrossRef]
- Tsukamoto, S.; Takeuchi, M.; Kawaguchi, T.; Togasaki, E.; Yamazaki, A.; Sugita, Y.; Muto, T.; Sakai, S.; Takeda, Y.; Ohwada, C.; et al. Tetraspanin CD9 modulates ADAM17-mediated shedding of LR11 in leukocytes. Exp. Mol. Med. 2014, 46, e89. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Thery, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef]
- Andreu, Z.; Yanez-Mo, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef]
- Sung, B.H.; Weaver, A.M. Exosome secretion promotes chemotaxis of cancer cells. Cell Adhes. Migr. 2017, 11, 187–195. [Google Scholar] [CrossRef]
- Sung, B.H.; Ketova, T.; Hoshino, D.; Zijlstra, A.; Weaver, A.M. Directional cell movement through tissues is controlled by exosome secretion. Nat. Commun. 2015, 6, 7164. [Google Scholar] [CrossRef]
- Chanda, D.; Otoupalova, E.; Hough, K.P.; Locy, M.L.; Bernard, K.; Deshane, J.S.; Sanderson, R.D.; Mobley, J.A.; Thannickal, V.J. Fibronectin on the Surface of Extracellular Vesicles Mediates Fibroblast Invasion. Am. J. Respir. Cell Mol. Biol. 2019, 60, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Purushothaman, A.; Bandari, S.K.; Liu, J.; Mobley, J.A.; Brown, E.E.; Sanderson, R.D. Fibronectin on the Surface of Myeloma Cell-derived Exosomes Mediates Exosome-Cell Interactions. J. Biol. Chem. 2016, 291, 1652–1663. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, R.; Kemper, S.; Brigstock, D.R. Structural and Functional Characterization of Fibronectin in Extracellular Vesicles from Hepatocytes. Front. Cell Dev. Biol. 2021, 9, 640667. [Google Scholar] [CrossRef] [PubMed]
- Gooz, P.; Dang, Y.; Higashiyama, S.; Twal, W.O.; Haycraft, C.J.; Gooz, M. A disintegrin and metalloenzyme (ADAM) 17 activation is regulated by alpha5beta1 integrin in kidney mesangial cells. PLoS ONE 2012, 7, e33350. [Google Scholar] [CrossRef] [PubMed]
- Trad, A.; Riese, M.; Shomali, M.; Hedeman, N.; Effenberger, T.; Grotzinger, J.; Lorenzen, I. The disintegrin domain of ADAM17 antagonises fibroblastcarcinoma cell interactions. Int. J. Oncol. 2013, 42, 1793–1800. [Google Scholar] [CrossRef] [PubMed]
- Roberts-Dalton, H.D.; Cocks, A.; Falcon-Perez, J.M.; Sayers, E.J.; Webber, J.P.; Watson, P.; Clayton, A.; Jones, A.T. Fluorescence labelling of extracellular vesicles using a novel thiol-based strategy for quantitative analysis of cellular delivery and intracellular traffic. Nanoscale 2017, 9, 13693–13706. [Google Scholar] [CrossRef]
- Toribio, V.; Morales, S.; Lopez-Martin, S.; Cardenes, B.; Cabanas, C.; Yanez-Mo, M. Development of a quantitative method to measure EV uptake. Sci. Rep. 2019, 9, 10522. [Google Scholar] [CrossRef] [PubMed]




