The large production of non-degradable petrol-based plastics has become a major global issue due to its environmental pollution. Biopolymers produced by microorganisms such as polyhydroxyalkanoates (PHAs) are gaining potential as a sustainable alternative, but the high cost associated to their industrial production has been a limiting factor. Post-transcriptional regulation is a key step to control gene expression in changing environments and has been reported to play a major role in numerous cellular processes. However, limited reports are available concerning the regulation of PHA accumulation in bacteria, and many essential regulatory factors still need to be identified. Here, we review studies where the synthesis of PHA has been reported to be regulated at the post-transcriptional level, and we analyze the RNA-mediated networks involved. Finally, we discuss the forthcoming research on riboregulation, synthetic and metabolic engineering which could lead to improved strategies for PHAs synthesis in industrial production, thereby reducing the costs currently associated with this procedure.
1. Introduction
The word “bioplastics” has commonly been used to make a distinction from petrochemical polymers, which is partially misleading, since not all types of bioplastics are bio-based and biodegradable
[1][2] (
Figure 1A). Some bioplastics are biodegradable but fully fossil-based. Their chemical structure can be degraded in a slow process catalyzed by enzymes of some aerobic and anaerobic microorganisms that are widely distributed in various ecosystems. However, they are not biodegradable in animal bodies and sometimes they remain in marine waters
[1][3] (
Figure 1A, bottom-right). Others are bio-based but chemically identical to their fossil counterparts, so they are not biodegradable
[1][4] (
Figure 1A, upper-left).
Only bio-based and biodegradable bioplastics are more ecologically friendly and serve as the best substitute for conventional plastics (
Figure 1A, upper-right). Among them, one of the most promising class of bioplastics are the bacterial polyesters polyhydroxyalkanoates (PHAs), which are produced through industrial bacterial fermentation of sugar or lipids by numerous Gram-positive and Gram-negative bacteria
[1][3]. Inside the cells, PHAs molecules aggregate to form water-insoluble granules, the carbonosomes, which are intracellular reserves of energy during starvation
[5][6] (
Figure 1B). In carbonosomes there is a constant cycle of synthesis and degradation, and this bidirectional process is a great advantage in the adaptation to rapid changes in the environment
[7][8]. During the last few years, PHAs are being proclaimed as the best alternative to fossil-based plastic due to their good balance between biodegradability rate, material properties that range from thermoplastics to elastomers, and the possibility to be processed into different final products
[9][10][11]. However, production costs of PHAs are still too high when compared to the synthetic plastics
[12][13]. Although they have not yet reached industrial scale, in the last decade a more cost-effective processes for the production of PHA have been developed based on the use of wastes, industrial products and less energy-demanding approaches
[14][15]. Once the process scale constraints are overcome, PHA will become more competitive and replace the synthetic plastics in many applications.
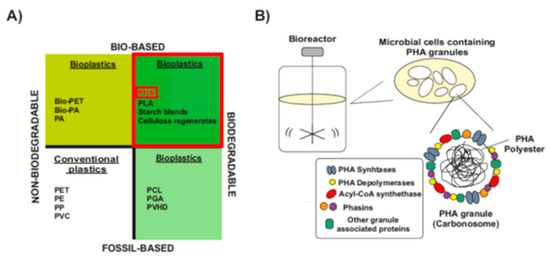
Figure 1. Material coordinate system of plastics. (
A) Type of plastics. Division of plastics into four groups, according to their biodegradability and biological origin. Upper-right, PHA: polyhydroxyalkanoates-are biodegradable polymers naturally produced by numerous microorganisms (Modified after
[1][2]).
(B) PHAs: bio-based biodegradable plastics. When a carbon substrate is present in excess, in parallel to depletion of other nutrients essential for biomass formation, PHAs are stored in the form of cytoplasmic spherical inclusions. These PHA granules are multi-complexes usually called “carbonosomes”. They contain a hydrophobic core surrounded by PHA granule-associated proteins, such as PHA synthase, PHA depolymerases, regulatory and structural proteins (Modified after
[7][16]).
Types and Chemical Structure of PHAs Polymers
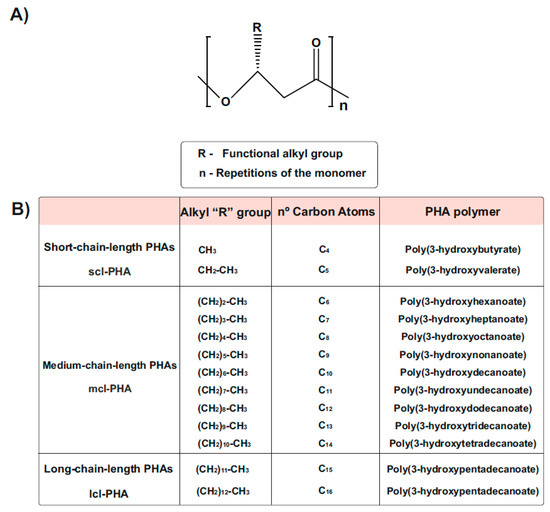
PHA generally consists of (R)-hydroxy fatty acid monomer units, which contain an alkyl side chain R group that varies in carbon length from methyl (C1)
[17][18] (
Figure 2). These polymers are usually divided into three different types, according to the number of carbons in the monomeric subunits
[18]. Short-chain-length PHA (scl-PHA) polymers are composed of monomers containing 3 to 5 carbon atoms, whereas medium-chain-length PHA (mcl-PHA) polymers are composed of monomers containing 6 to 14 carbon atoms. The third type are the long-chain-length PHAs (lcl-PHA), with a minimum 15 carbons
[17][18] (
Figure 2B). Their chemical properties are different and depend on the bacterial host and the fermentation conditions used for their production, making them suitable for different purposes. Scl-PHAs are highly crystalline, which makes them relatively stiff and brittle
[17][19]. However, polymers with a greater number of carbons are more flexible and elastic, resulting in increased research interests
[20].
Figure 2. Chemical Structure of polyhydroxyalkanoates (PHAs). (
A) General structure of a PHA molecule. The ’R’ functional group represents the alkyl side chain, and the number of repetitions of the monomeric unit is given by ‘n’. (
B) PHAs can be classified as scl-PHAs, mcl-PHAs, and lcl-PHAs, depending on the carbon numbers in the monomeric constituents (modified after
[18]).
PHAs are classified into homopolyesters, with only one variety of monomer, and heteropolyesters, which can be subdivided into copolyesters (monomers differing in either backbones or side chains) and terpolyesters (different side chains and backbones)
[16][21]. The so-called polyhydroxybutyrate (PHB) is one of the most common homopolymer PHA and best studied scl-PHA, containing the shortest possible side chain with only one methyl group
[22][23] (
Figure 2B).
Natural PHA Producers and Engineering of Non-PHA Producers
Although the list of natural PHA producers is large and includes extremophile bacteria, mainly Gram-negative species have been explored for their capacity to synthesize PHAs. Among this list, the most known are:
Cupriavidus necator (previously
Ralstonia eutropha),
Azotobacter vinelandii, and
Burkholderia spp., as scl-PHA producers
[18][24][25];
Pseudomonas strains (especially
Pseudomonas putida), as mainly mcl-PHA accumulators, while some strains are able to produce scl-mcl-PHA co-polymers
[26][20][27].
In the last few years, new knowledge was gained about biosynthetic pathways (largely confined to Acetyl-CoA precursors) and the enzymes involved in PHAs accumulation
[22][28]. Neverth
PHA Composition and Preferred Carbon Source
To produce PHAs, bacteria can use different carbon sources as substrate such as saccharides, fatty acids, alcohols, or gases
[18][29]. Generally, different bacteria have preference for one of them depending on the metabolic pathways they harbor, so the metabolic routes in which those substrates are integrated are different, as well as the final product composition
[18][22][30].
The genes (
pha) that regulate the synthesis and degradation of PHA at the transcriptional level are widely known among the prokaryotes. In the extensively studied
P. putida, the genetic organization of the
pha genes integrates a very conserved
pha cluster composed by two synthases (
phaC1 and
phaC2) responsible for the PHA synthesis; a depolymerase (
phaZ) encoding for the PHA mobilization; the transcriptional regulator (
phaD); and the regulatory and functional phasins (
phaF and
phaI)
[31][8][32]. In the last few years, new knowledge has been deciphered about the PHAs synthesis and degradation in pseudomonads and other organisms
[20][33][34][35]. However, the molecular regulation at the post-transcriptional level of PHA synthesis is still unclear and needs further investigation.
2. RNA-Mediated Control in Native Synthesis of PHAs
2.1. The Expanding RNA World: Non-Coding Bacterial RNome
For years, it was considered that the expression of the bacterial genome resulted in three large groups of RNA molecules: mRNA, which contains open reading frames that translate into proteins, and two more types of RNA, the ribosomal rRNA and transfer tRNA, which are essential for protein biosynthesis carried out by ribosomes. Therefore, regulation of gene expression was exclusively associated with the activity of protein regulators [36][37]. However, post-genomic research is revealing an unprecedented high abundance and diversity of untranslated small RNA molecules (50–350 nt of average length) called sRNAs or non-coding RNAs, expanding the total of RNA species that together constitute the bacterial “RNome” [38][39]. These sRNAs are commonly encoded by single transcriptional units between open reading frames (ORFs) and although do not translate to protein, play very important roles in the gene regulation of diverse physiological processes at the post-transcriptional level [40][38][41].
These riboregulators are deeply conserved in prokaryotes and adjust gene expression in response to specific environmental or physiological signals, facilitating adaptation to diverse environmental stimuli. This is especially important to allow the cell to profit from transiently available nutrients [38][42].
Depending on their genomic location relative to the mRNA targets that they regulate, sRNAs are classified as cis- or trans-encoded. The latter constitute a majority group and are expressed from intergenic regions (IGRs), generally far from the target messenger counterparts [43][44][45].
2.1.1. RNA-Binding Proteins and Regulatory Networks
Some regulatory RBPs can act as chaperones by facilitating the intermolecular base pairing between sRNAs and mRNAs [46][47]. One of the best characterized RBP is Hfq that also exerts a central role in post-transcriptional gene regulation, as evidenced by the pleiotropic effect of the inactivation of the hfq gene in many Gram-negative bacteria [48][49][50][51].
As most sRNAs have typical bacterial Rho-independent terminators that usually contain a poly-U 3′-terminus, Hfq can interact with the terminators and influence sRNA stability [52]. The distal face has strong affinity for A-rich sequences of mRNAs [53][54].
Hfq also offers a scaffold for the interaction with several other proteins [51], e.g., Crc, which is involved in catabolite repression control in some Pseudomonas sp. [46][51][55][56].
2.1.2. Post-Transcriptional Regulation by Ribonucleases
Ribonucleases are enzymes that have been widely described to play important and even essential roles. As reviewed in
[48][57][58], RNases are essential participants during the post-transcriptional regulation and as key modulators of RNA decay. In general, sRNAs are mainly degraded by RNase E and PNPase, or by RNase III, if the sRNA is hybridized to an mRNA target
[48].
RNase E is the major bacterial endoribonuclease and cleaves single-stranded regions of structured RNAs with preference for 5′-ends and AU-rich sequences
[59]. This ribonuclease is the main enzyme forming the degradosome, a ribonucleoprotein (RNP) complex involved in the decay of many RNAs
[58][60].
Another RNase involved in post-transcriptional regulation by bacterial sRNAs and through the decay of some mRNAs is the double-strand specific RNase III
[61][62]. It is a highly conserved enzyme specific for double-stranded RNAs which shows preference for continuous RNA duplexes of 20–40 bps. Perfect antisense/sense RNA duplexes formed in sRNA–mRNA interactions constitute an optimal substrate for this enzyme
[63].
YbeY is an additional RNase that has been recently proposed to be required for the sRNA-mediated post-transcriptional silencing of prokaryotic genes. This endoribonuclease cleaves double-stranded RNA and could have catalytic and/or Hfq-like protective functions essential for RNA metabolism and small RNA (sRNA)-mediated regulation
[64].
2.2. Post-Transcriptional Regulation of sRNAs and Their Implications for Microbial PHAs Synthesis in Different Microorganisms
2.2.1. MmgR sRNA Is a Negative Regulator of PHB Accumulation in Sinorhizobium meliloti
The
S. meliloti genome is distributed in three replicons (3.65-Mb chromosome, the megaplasmids 1.35-Mb pSymA, and 1.68-Mb pSymB) and encodes more than 500 sRNA candidates
[49][65]. However, with only a few exceptions, the regulatory targets and mechanism of action of this repertoire of sRNAs are still unknown
[66][67][68][69]. The
S. meliloti trans-encoded sRNA, MmgR (standing for Makes more granules Regulator) is an Hfq-dependent sRNA
[49][70], transcribed from the chromosome as a 77-nt RNA
[67][71][72]. It is highly conserved in α-proteobacteria, as a member of the αr8 RNA family, and has been explored for its regulatory function only in
S. meliloti [67][72]. Lagares et al.
[67] found that MmgR is a negative regulator of PHB accumulation since the deletion of an internal conserved core of the sRNA gene resulted in larger cells containing 20% higher amounts of PHB (Figure 5 in ref.
[67]). Further, the
mmgR expression was described to be modulated by the availability of N existing in the growth medium
[67][73].
Phasins mediate stabilization of the granule–cytoplasm interphase
[74]. In agreement with this, quantitative reverse transcription-PCR (qRT-PCR) and proteomic profiling enhanced the accumulation of PhaP1 and PhaP2 proteins in the
mmgRΔ33–51 mutant without affecting their mRNAs levels. These decoupling results evidenced the post-transcriptional negative regulation that the MmgR sRNA carries out, in a direct or indirect manner, on the
phaP1 and
phaP2 mRNAs in
S. meliloti [67].
The promoter activity of
mmgR is controlled by the quality and/or amount of the available N source, reaching the highest intracellular level with nitrate as the N source or upon starvation of the organic N sources
[67][75]. The expression of MmgR was mainly regulated at the transcriptional level by at least the N and C metabolism master regulators NtrC and AniA, respectively. This regulation relies on a conserved dyadic motif located within the −35 and −10 boxes of the
mmgR promoter, and results in positive control of gene expression by the C:N molar ratio in the growth medium, upon N depletion. On the other hand, the global carbon flux regulator, AniA (PhaR), negatively controls the sRNA expression, assuming a consistent negative feedback loop on phasin and
phaZ genes since the MmgR sRNA down-regulates PhaP1 and PhaP2 protein levels
[67][73].
2.2.2. Post-Transcriptional Control of PhbR as Key Step during PHB Production in Azotobacter vinelandii
The main regulatory mechanism leading to the accumulation of PHB in this organism involves the phbBAC operon, which encodes for key enzymes of the PHB biosynthesis pathway. This operon is in turn controlled by the transcriptional activator PhbR and the sigma factor RpoS [76][77]. Interestingly, PhbR expression has been reported to be post-transcriptionally controlled by the two-component GacS–GacA global regulator [78]. This system (global antibiotic and cyanide control) belongs to the Gac–Rsm cascade and is involved in the regulation of many cellular processes in numerous bacterial organisms, as reviewed by Lapouge et al. [79].
Hernandez-Eligio et al.
[78], moreover, revealed that the RsmA protein targets and causes instability on both,
phbR and
phbB mRNAs. Further analysis uncovered that the mutation in the
rsmA gene generates an increase in the translation of
phbR/phbB, whereas a strong reduction in their activity was observed in the
gacA mutant, without determining whether the mutation affects the translation of
phbB. Taken together, these results confirmed that the Gac–Rsm system controls
phbR expression at the post-transcriptional level in this strain, while it could not be established for the regulation on
phbB [78][80]. The model shown in Figure 7 of Hernandez-Eligio et al.
[78] properly summarizes this regulatory control of PhbR by the Gac–Rsm cascade. It is also possible to consider RsmA (CsrA) as the central component of the system. Therefore, additional research (on the
phbB regulation and the interaction of the sRNAs with the RsmA protein) is needed for the further understanding of the control of PHB through this regulatory cascade in
A. vinelandii UW136
[78][81].
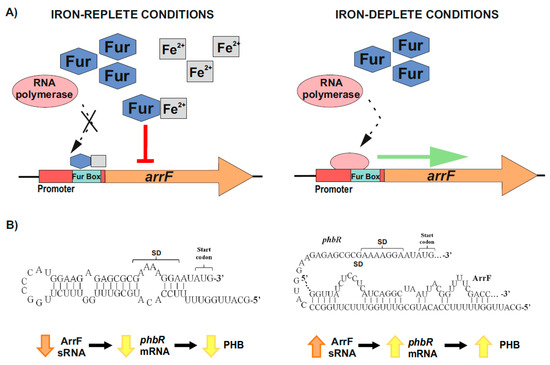
On the other hand, and over the past few years, it has been revealed that bacterial iron-regulated sRNAs have important modulating roles (e.g., in iron homeostasis) according to the levels of this essential and potentially toxic micronutrient
[82][83][84]. The genes that encode for these small RNAs hold in their promoter regions the conserved Fur or iron boxes, which function as binding sites of the ferric uptake repressor (Fur)
[80][83][84][85][86]. Under iron-replete conditions, a Fur–Fe
2+ complex is formed, which binds to the iron boxes of the sRNAs-coding genes involved in iron homeostasis, and represses their transcription. On the contrary, when iron is scarce, RNA polymerase is able to access the promoters of these genes, resulting in their transcription
[80][83][84][85][86] (
Figure 3A). In
A. vinelandii, iron regulates the accumulation of PHB through one of these sRNAs, ArrF, while the mechanism seems to vary between different genetic backgrounds
[82][87][80][88]. Muriel-Millan et al.
[82] reported that, under iron limitation, the ArrF sRNA acts as a positive post-transcriptional regulator of the
phbR gene. In the proposed mechanism, the
phbR mRNA forms an inhibitory hairpin around the Shine–Dalgarno (SD) sequence in the ribosomal binding site (RBS), thereby preventing initiation of translation. When the levels of ArrF rise, this antisense sRNA binds to a complementary target sequence within the 5′ UTR of the
phbR mRNA
[82][80][86][88] (
Figure 3B). In strain UW136, this interaction releases the inhibitory hairpin structure in the mRNA, unblocking the SD and allowing translation, which in turn increases PHB production
[82]) (
Figure 3B). However, in strain KCTC 23243 (whose wild-type is able to synthetize only small PHB quantities), this interaction results in a downregulation of
phbR gene expression and therefore less accumulation of PHB
[88].
Figure 3. Iron-mediated regulation of PHB accumulation through the ArrF sRNA in
A. vinelandii. (
A) Upper left panel, under iron-replete conditions, Fur–Fe
2+ complexes are established and repress
arrF genes transcription by binding the Fur boxes in their promoters; Upper right panel, under iron-deplete conditions, RNA polymerase accesses the
arrF promoter with its subsequent transcription. (
B) The mechanism through which ArrF sRNA is involved in the PHB accumulation differs between different genetic backgrounds. Left panel, in
A. vinelandii strain UW136, a predicted occluding hairpin around the
phbR 5′-UTR could prevent its translation; Right panel, the targeting of
phbR 5′-UTR by the ArrF sRNA could make the RBS available for translation, resulting in turn in an increase in PHB accumulation (modified after
[82]).
2.2.3. Global Post-Transcriptional Regulatory Protein Crc as Main Target of sRNAs CrcZ and CrcY in Pseudomonas putida
The catabolite repression control protein (Crc) plays a key role in the CCR process, impeding the expression of genes involved in the synthesis of catabolic enzymes for the use of non-preferred carbon sources in pseudomonads [89][90][91][92]. This global regulator recognizes AANAANAA sequences in the genome called catabolite activity (CA) motif, located near the Shine–Dalgarno sequence of target mRNAs, and with a function in translation inhibition [93][55][89][94][92][95]. This process is in cooperation with the distal face of the protein Hfq, which is required by Crc to bind the mRNA motif through the formation of a stable ribonucleoprotein complex at the targets [96][97][95][98][99]. In Figure 9, Moreno et al. [89] summarizes the procedure by which the action of this complex is modulated and antagonized by two small RNAs (CrcZ and CrcY) in P. putida. The levels of both sRNAs significantly increase when bacteria grow with a non-preferred carbon source or have reached stationary growth phase. These sRNAs sequester one or both of the Crc/Hfq proteins, therefore decreasing the CCR, and allowing translation of the target mRNAs with A-rich motifs involved in the transport and/or assimilation of compounds [100][89][101][96][97][95][98][99]. During the formation of this multilayered and complex Hfq/Crc/CrcZ-CrcY regulatory system, the Crc–Hfq complex protects the sRNAs from ribonucleases by increasing their stability [48][97][102]. These sRNAs are mainly transcribed from σ54/RpoN-dependent promoters (PcrcZ and PcrcY) regulated by the two-component sensor-regulator system CbrA–CbrB (mainly CrcZ) together with other protein factors [93][100][89][101][96][97][92]. In this regulatory complex, each component affects either the transcription or the stability of the other components, e.g., the activity of the sRNA promoters relies on the type of carbon source and carbon/nitrogen (C/N) ratio. In turn, this promotes that the cellular metabolism adopts distinct pathways that allow the cell to adapt its requirements for energy and molecular biosynthesis [93][89][101][97][92].
2.2.4. Post-Transcriptional Control of phaC1 Synthase as a Key Aspect along PHA Synthesis in P. putida CA-3
To further analyze the role of this two component system in
Pseudomonas putida CA-3, Ryan et al.
[103] performed a screening of a random mini-Tn5 mutagenesis of its genome, in which 44 mutants were identified with a reduced PHA accumulation phenotype. After the characterization of one of these mutants (PHA45A) that has a disruption of the
gacS gene, it was ultimately concluded that this sensor kinase is directly related with the post-transcriptional regulation of PHA synthesis in this strain
[103][80]. To reach this conclusion, first the identification of Gac–Rsm cascade gene homologues in
P. putida CA-3 was accomplished, followed by the evaluation of their genetic expression in both wild-type and
gacS mutant backgrounds, under PHA accumulation conditions. However, and in contradiction with the model of transcriptional regulation within the cascade known in other pseudomonads, the transcription of the sRNAs RsmY and RsmZ (previously identified in strain CA-3), was not affected in the PHA45A mutant. Similarly, the expression of the PHA biosynthetic genes
phaC1 polymerase and
phaG-encoded ACP-CoA transacylase
[104], presented similar transcript levels in both genetic backgrounds, analyzed under the same conditions
[103][80].
Despite these results in the expression of the analyzed genes, evidence exists that the
gacS disruption in the PHA45A mutant of
P. putida CA-3 inhibits PHA accumulation. Therefore, the possible regulation of the PHA synthesis at a post-transcriptional level was investigated
[103]. Subsequently, the protein profile of the
gacS mutant was evaluated together with an already characterized
phaC1-disrupted mutant, whose protein had been previously reported to be essential for PHA accumulation in this strain
[104]. Both strains exhibited an absence of protein at the expected ∼62 kDa band, compared with the wild-type protein profile. Hence, Ryan et al.
[103] concluded that the post-transcriptional regulation of the PhaC1 PHA synthase was the key step in the GacS regulatory cascade along PHA synthesis in
P. putida CA-3. This unusual procedure could involve other regulatory elements controlling RsmY and RsmZ sRNAs for PHA synthesis in
P. putida CA-3, which would need further research to be fully understood
[31][103][80].
3. Conclusions and Perspectives
PHAs are polyesters synthesized and biodegraded by microorganisms, which are produced from large accessible renewable resources and have potential use for numerous applications. However, detailed understanding and subsequent optimization of their production and purification are still mandatory to reduce their production costs
[10][22][25][105].
3.1. Role of Post-Transcriptional Regulation during the Native Synthesis of PHAs
Free-living bacteria often need to develop flexible and versatile metabolic and regulatory networks to adapt to fast fluctuations in nutrient availability. Therefore, the destiny of C aims to maximize bacterial fitness and safety
[99]. Phylogenetic analysis of the ability of bacteria and archaea to synthesize PHAs has revealed extensive horizontal gene transfer events of the genes and corresponding transcriptional regulators involved in the accumulation of these polymers
[67][106]. However, in the vast majority of cases, their post-transcriptional regulation still remains unknown.
Riboregulation has a major role in the fine-tuning of multiple bacterial processes and is important to rapidly adjust cell growth in response to environmental changes
[42]. sRNAs are non-translated small RNA molecules that are very important in the control of gene expression that usually silence their targets
[41]. Ribonucleases are the enzymes that process and degrade all types of RNA and it is known that the RNA chaperone Hfq can protect RNA from the action of ribonucleases
[48][107]. As shown in this review, the PHAs synthesis is also adjusted, directly or indirectly, through post-transcriptional regulation exerted by different kinds of RNAs molecules
[93][103][82][67][78]. Although nothing has been published about the implication of RNases in the control of PHAs synthesis, they are expected to play an important role based on their marked importance in controlling other regulators and processes
[64][57][108].
3.2. Controlling PHAs Production in Bacteria via Synthetic Small Non-Coding RNAs
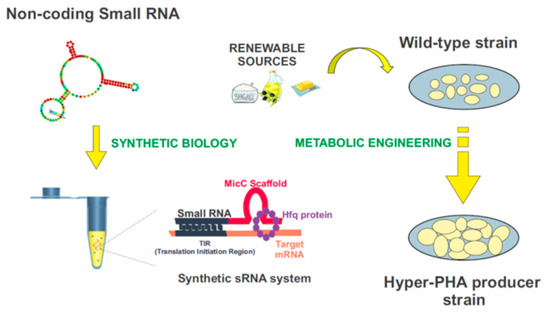
Synthetic biology is a compelling and expanding interdisciplinary research field which intends to provide a systematic framework for the design and construction of biological systems. It relies on the application of logical engineering principles to program or reprogram cellular functions at a genetic and metabolic level (
Figure 4)
[109]. One of the most important endeavors in contemporary synthetic biology is the search for optimal genomic chassis for industrial applications
[110][111]. With this idea in mind, there has been a great effort to develop customizable regulators using genetic tools such as the CRISPR/Cas system, TALEs, and sRNAs
[112], which would enable the precise control of gene expression, aiming to attain the desired functional outputs. Driven by the widespread role of post-transcriptional regulation in natural systems, the attention paid to RNA regulators is increasing
[113]. Recent advances in nucleic acid engineering encourage the design of RNA components as building blocks in the construction of synthetic biological systems, mainly due to the plasticity of these molecules to interact with a myriad of proteins, metabolites, and other nucleic acids
[114]. Synthetic RNA regulators display a wide range of programmable functions, offering important advantages over other protein-based mechanisms
[108]. Among them, synthetic small non-coding RNAs (synthetic sRNAs) emerge as promising components to fine-tune gene expression. These customizable RNA regulators can be rationally designed to target different mRNAs, modulating their expression by altering their target-binding sequences (
Figure 4)
[31][108][115][116][117].
Figure 4. Biotechnological domestication of microorganisms to improve PHA synthesis. Synthetic biology and metabolic engineering strategies are used in the bacteria “domestication”. Customizable synthetic small RNAs could be rationally designed using the MicC scaffold to target genes of interest, and improve the industrial production of PHAs.
Improving the quality and reducing the costs in industrial production of PHAs is a matter of pressing importance. In the future, synthetic sRNAs could be used to domesticate bacteria throughout the modulation of their genetic expression, in particular on the enzymes involved in the PHAs synthesis. The construction of these customized sRNA systems could be used for this purpose, in combination with the use and further development of plasmid genetic tools, such as the SEVA plasmids, for the modulation of genes-of-interest
[31][108][115][116][117][118][119].
Figure 4 exemplifies this process. The MicC scaffold can be used to design tailor-made sRNAs that target the genes of interest, with the help of the Hfq protein. More details about this synthetic sRNA system can be found in
[108][115][116][117].
It is important to continue investing the biotechnological domestication of microorganisms using synthetic biology and metabolic engineering to implement the portfolio of PHAs and improve strategies to lower the costs in industrial production
[31][120]. In this review, we have indicated many examples of how post-transcriptional control can be an instrumental tool for the regulation of polyhydroxyalkanoates synthesis.