Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Amit Singh | + 3207 word(s) | 3207 | 2021-08-18 09:44:46 | | | |
| 2 | Peter Tang | Meta information modification | 3207 | 2021-08-26 04:00:09 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Singh, A.; Cabral, C.; Gupta, A. Linkage of Dysbiosis to Diseases. Encyclopedia. Available online: https://encyclopedia.pub/entry/13548 (accessed on 07 February 2026).
Singh A, Cabral C, Gupta A. Linkage of Dysbiosis to Diseases. Encyclopedia. Available at: https://encyclopedia.pub/entry/13548. Accessed February 07, 2026.
Singh, Amit, Célia Cabral, Ashutosh Gupta. "Linkage of Dysbiosis to Diseases" Encyclopedia, https://encyclopedia.pub/entry/13548 (accessed February 07, 2026).
Singh, A., Cabral, C., & Gupta, A. (2021, August 25). Linkage of Dysbiosis to Diseases. In Encyclopedia. https://encyclopedia.pub/entry/13548
Singh, Amit, et al. "Linkage of Dysbiosis to Diseases." Encyclopedia. Web. 25 August, 2021.
Copy Citation
The human intestine contains an intricate ecological community of dwelling bacteria, referred as gut microbiota (GM), which plays a pivotal role in host homeostasis. Multiple factors could interfere with this delicate balance, including genetics, age, antibiotics, as well as environmental factors, particularly diet, thus causing a disruption of microbiota equilibrium (dysbiosis). Growing evidences support the involvement of GM dysbiosis in gastrointestinal (GI) and extra-intestinal cardiometabolic diseases, namely obesity and diabetes.
dietary polyphenols
gut microbiota
dysbiosis
gastrointestinal diseases
metabolic disorders
delivery systems
1. Introduction
Hippocrates has been cited as saying “death sits in the bowels” and “bad digestion is the root of all evil” in around 400 B.C., suggesting the important role of the human intestine in health and disease. Gut microbiota (GM) is the collective community of microorganisms living in the gastrointestinal (GI) tract. Approximately 100 trillions of microorganisms, consisting mainly of bacteria, inhabits in the human GI tract [1]. Viruses, protozoa and eukaryotic organisms, such as fungi, are also present in a small number. In the adult GI tract about 90% of the bacteria fit in the phyla Bacteroidetes (Gram-negative) and Firmicutes (Gram-positive), while other phyla are present in much lower abundance, such as Actinobacteria (Gram-positive), namely Bifidobacterium, Proteobacteria (Gram-negative) and Verrucomicrobia (Gram-negative), namely Akkermansia muciniphila (Gram-negative) [1][2].
In spite of the fact that people have several hundreds of microbial species inside their gut, newer findings obtained by the Human Microbiome Project and other relevant studies demonstrate that microbial composition is highly variable between individuals [3]. Bacterial colonization starts in utero and GM composition changes throughout the entire life, but the main changes, in number and in diversity, occur during the breast-feeding period and at the beginning of solid food ingestion. The number, type and function of microorganisms differ throughout the GI tract but the bulk is found inside the large intestine, participating in fermentation of undigested food components, particularly carbohydrates and fibers, among other relevant functions. The main roles of GM in humans are depicted in Figure 1. Apart from affording protection against enteropathogens and absorb nutrients from our diet, GM produces several bioactive compounds, some of which are beneficial to health, namely vitamins and some short chain fatty acids (SCFAs), while others are deleterious, such as some metabolites of degradation of amino acids. In addition, host immune defenses, in particular the mucus barrier, are important to protect tissues against harmful effects of some bacteria.
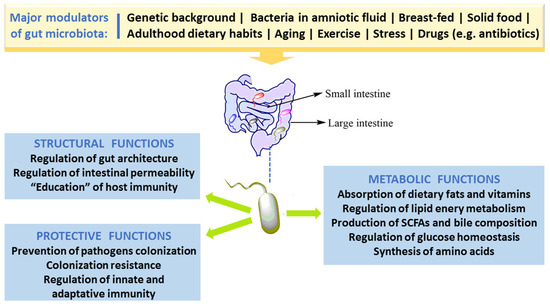
Figure 1. Major factors that modulate gut microbiota and key roles in humans.
Factors responsible for an impaired GM composition and/or function, called dysbiosis, include age, diet and lack of exercise, stress, drugs and xenobiotics [1][4][5]. There is increasing evidence supporting an association between dysbiosis and diseases, including those of the GI tract, such as inflammatory bowel disease (IBD), ulcerative colitis (UC), Crohn disease (CD) and colorectal cancer (CRC) [6][7][8], as well as some extra-intestinal metabolic disorders, including obesity, diabetes and its macro- and microvascular complications [9][10][11]. Hence, researchers all over the world are searching for therapeutic or nutraceutical interventions able to produce a healthy GM equilibrium, eliminating the harmful bacteria (or pathobionts) without affecting the beneficial ones (symbionts).
Dietary polyphenols are natural compounds present in many foods and beverages, namely in fruits, vegetables, cereals, tea, coffee, and wine, among others. Several preclinical and clinical studies have shown their antioxidant, anti-inflammatory, anti-diabetic, anti-cancer, neuroprotective, and anti-adipogenic properties, suggesting a link between polyphenol-rich food consumption and reduction in the incidence of numerous chronic disorders, highlighting them as good candidates for therapeutic/nutraceutical agents [12][13][14][15]. However, inside the human body, the chemical structure of the majority of polyphenols is received as a xenobiotic and, thus, the bioavailability of these compounds is highly reduced when compared to that of macro- and micro-nutrients [16]. Because of poor absorption, they are retained in the intestine for longer time where they can promote beneficial effect, namely by affecting the GM community [17]. The impact of dietary polyphenols on gut ecology and the mechanism underlying the putative beneficial effects on GI and extra-intestinal diseases have been depicted during the last decade [18].
2. Gut Microbiota Dysbiosis and Disease
2.1. Gut Morphology and Healthy Microbiota Composition, Diversity and Functions
The human GI tract, with 250–400 m2 surface area, is one of the chief associations between the host body, external environmental factors and internal antigens. During average lifespan, about sixty tonnes of food and a huge number of external microbes traverses the GI tract, jeopardizing gut integrity. The set of gut microbes- bacteria, archaea, viruses (mainly phages), eukaryotes (mainly yeasts), and other microbial species-, collectively referred as gut microbiota, has jointly developed with the host during millions of years, forming a complex and symbiotic connection [19].
Among the large population of microbial species, about 1014 cells inhabit the human GI tract. The composition of GM varies along the GI tract in accordance with the morphological and physiological features of digestive system region. The concentration considerably rises from the proximal to the distal gut, together with an enrichment in anaerobes [1][2]. GM composition in the initial proximal portion of the small intestine, predominantly in the duodenum, is identical to that of stomach, given the acidic conditions that results from the chyme of the stomach and biliary and pancreatic secretions. The diversity and number of bacteria increases in the distal portion, from duodenum to ileum, accompanying the gradual increase of pH. In this portion, predominate Lactobacillus and Clostridium species of the Firmicutes phyla, Escherichia coli of the Proteobacteria phyla, as well as Bacteroidetes and gram-negative facultative anaerobes [1][2]. The conditions in large intestine, namely in the colon, with a more beneficial nutritional milieu and pH (5.7–6.8), favor bacterial growth, allowing a more diverse, dense and complex microbial community, essentially composed of obligate anaerobes that survive at low concentrations of oxygen. In the colon, the predominant bacteria are Ruminococcus, Lactobacillus and Clostridium species of the Firmicutes phyla, and Bacteroides and Prevotella of the Bacteroidetes phyla. There are other phyla present in the adult GM, thought in lower abundance, including Actinobacteria, Fusobacteria, Proteobacteria and Verrucomicrobia, as well as some facultative anaerobic bacteria [20][21]. Bacterial composition and functions also varies between the intestinal lumen and the mucus layer of the intestinal mucosa, with a different proportion of anaerobic and aerobic species [1][2][22].
Several recent pieces of evidence support the idea that, against what was previously thought, the microbial colonization of humans starts in utero from maternal commensal microbes, as suggested by the bacteria found in the amniotic fluid, maternal placenta, umbilical cord blood and in meconium (the first “faeces-like excretion”) [23][24][25].
GM phylogenetic diversity increases with growth and development of the host; around 2–3 years age, a complex and stable community of microorganisms is formed. Based on different methods of molecular profiling, it is predictable that about one thousand species of bacteria inhabit the gut, mainly anaerobes, including Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria. However, the two predominantly phyla Bacteroidetes and Firmicutes constitute over 90% of all bacterial species in the intestine [26][27].
The healthy GM is stable and contributes to several important physiological host functions (Figure 1). In brief, GM exerts important (i) metabolic effects (co-metabolism), such as synthesis of vitamins and fermentation of carbohydrates, lipids, and proteins; (ii) structural defending properties against pathobionts by preserving integrity and regulating the permeability of gut barrier, thus contributing to host homeostasis; as well as (iii) “education” of host immunity [5]. The intestinal bacteria have ability to synthesize essential and nonessential amino acids. They can produce numerous vitamins and short-chain fatty acids (SCFAs) and are able to carry out biotransformation of bile. Moreover, GM can metabolize some complex oligosaccharides which escaped the digestion, namely several barely digestible polysaccharides, including cellulose, hemicellulose, resistant starches, gums and pectins, unabsorbed sugars and alcohols obtained from the diet. This helps in the retrieval of absorbable substances to the host and in turn the bacteria derive nutrients and energy for their growth and multiplication [5][28]. GM plays a vital role in host immune system development and maintenance. Many findings proved the crucial role of GM in the regulation of antigen presenting cells (APCs) development, namely B-cells, macrophages and dendritic cells, which have capability to protect the body from pathogens while being immuno-tolerant towards gut microbes [29]. Studies with germ-free mice have reported that symbiotic intestinal bacteria are indispensable for the growth and functioning of intestine-linked lymphoid tissues (namely Peyer’s patches and mesenteric lymph nodes) and specific lymphocytes [30]. Under normal conditions, GM/immune system interaction stimulates the generation of beneficial responses towards pathogens and the protection of the regulatory pathways associated with the tolerance preservation towards safe antigens. In the developed countries, factors like unwarranted usage of antibiotics and major changes of dietary habits might have influenced the selection of a microbial community composition that fails to create balanced immune responses. This could be ascribed towards the noteworthy increase in autoimmune and inflammatory disorders spanning the last few decades [31].
2.2. Factors Responsible for Gut Microbiota Alteration (Dysbiosis)
In the mid-20th century, Metchnikoff first coined the term “dysbiosis” to depict the changes in intestinal bacteria, suggesting a link with immune homeostasis impairment and development of intestinal disorders. In general, dysbiosis can be classified as (1) a decrease in number of symbionts; (2) an unwarranted growth of pathobionts; and (3) a loss of diversity. It has been reported that these 3 different types can co-exist, which is most often the case. Several factors are responsible for an impaired GM composition and/or function-dysbiosis-, including age, diet and lack of exercise, stress, drugs and xenobiotics [1][4][5].
Major GM alterations have been reported in early phase of life, particularly in the first years, when there is a marked increase in numbers and diversity. The maturation and evolution of the human gut microbes is an example of ecological succession [32][33]. After an initial stage of massive new colonization, when the microbial composition is highly variable between individuals, GM undergo successive changes in composition and function until a stable climax community is established. However, major differences have been reported between adult populations of distinct geographic World regions, which might be explained by a diversity of factors, including both genetic and environmental, particularly dietary patterns, hygienic conditions, as well as use of antibiotics and other drugs [34][35]. Furthermore, aging also affects GM composition, changing the number and particularly the diversity of bacteria, with a decrease in the Firmicutes to Bacteroides (F/B) ratio, which might be partially explained by modifications caused by an altered immune-inflammatory milieu–the so called immunosenescence [10]. Further causes of aging-related altered GM are the deterioration of physical health, including loss of dentition, impaired salivary function, digestion and transit time in the GI tract, as well as abnormal dietary nutrients resulting in malnutrition [36][37].
Diet is in fact a chief GM modulator, affecting both the composition and functions, thus contributing to maintain health or to favor disease states [38]. For instance, breast milk contains certain oligosaccharides that cause proliferation of Lactobacillus and Bifidobacterium that are predominant in the infant gut and could contribute to immune system development. Similarly, demographic dietary factors result in differential variation in the GM [39]. Rural Africa children, usually consuming plant polysaccharides, had lower faecal levels of Firmicutes and higher of Bacteroidetes (particularly Prevotella and Xylanibacter) when compared with Italian kids that present high amounts of Enterobacteriaceae (especially Shigella and Escherichia). Prevotella and Xylanibacter, in particular, can cause an increment of beneficial SCFAs as a result of cellulose and xylans degradation, suggesting an adaptation to take full advantage of energy extraction from fibre-rich diet. In fact, resistant starch in human diets promote increment of Ruminococcus bromii and Eubacterium rectale in faeces, which show a relationship with fibre fermentation [40]. Several evidences from preclinical models and from humans studies consistently show that high-fat and/or high-sugar diets modifies the GM profile towards dysbiosis, while vegan diet, prebiotics (such as inulin) and/or probiotics causes beneficial effects on GM composition and function, accompanied by reduced adiposity, inflammatory molecules, including lipopolysaccharide (LPS), and other positive effects on metabolic hemostasis [4].
Some recent evidence points to the impact of artificial sweeteners and emulsifiers in GM. Artificial sweeteners, frequently used as sugar alternatives, have been “generally recognized as safe” (GRAS) by regulatory agencies. However, investigations have reported that some of them, including aspartame, sucralose and saccharin, may upset the balance and diversity of GM. In fact, rats orally treated with sucralose for 84 days showed increased gut levels of Clostridia, Bacteroides, and total aerobic bacteria, upregulated expression of bacterial pro-inflammatory genes, as well as disturbed faecal metabolites and augmented faecal pH [41]. Emulsifiers, which are food additives often present in processed food, have also been associated with altered GM in animals. Mice treated with carboxymethylcellulose and polysorbate-80, two commonly used emulsifiers, revealed decreased numbers of Bacteroidales and Verrucomicrobia together with increased amounts of Proteobacteria, which usually is linked with mucosal inflammation [42]. Altogether the previous examples reinforce the important role of diet in modulation of bacterial-derived metabolites than just influencing the microbiota community for short-term period [43].
Several non-dietary environmental factors are able to impact the human GM towards dysbiosis, including stress, smoking habits and lack of exercise practice, which might be due to, among other factors, the associated pro-oxidative and pro-inflammatory milieu [10]. In addition, medication is a major modulator of the gut ecology. By definition, antibiotics alter GM composition and functions [44][45] and the loss of colonization resistance is an earlier effect of antibiotics on the gut. This loss resulted in much easier rate of colonization by Salmonella following antibiotic treatment. Recent studies in mice reveal that antibiotics cause an increment of gut free sialic acid derived from the host, that can be further used by opportunistic pathogens to growth (namely by Salmonella typhimurium and Clostridium difficile) [46].
3. Linkage of Dysbiosis to Diseases
There are increasing evidences in favor of an association between dysbiosis and diseases, including those of the GI tract, such as IBD, UC, CD and CRC, but also some extra-intestinal metabolic disorders, such as obesity, diabetes and macro- and microvascular complications [8][9][10][11]. Figure 2 summarizes some of the main linkages of dysbiosis to GI and extra-intestinal metabolic and vascular diseases.
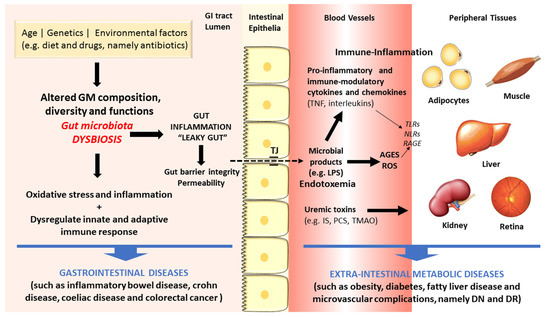
Figure 2. Schematic diagram of some of the main factors and pathways linking dysbiosis to gastrointestinal and extra-intestinal metabolic and vascular diseases.
The gut microbes act in tandem with the defense and immune systems of the host to protect against colonization and invasion of pathogens [21]. Recently, a remarkable amount of experimental data has firmly advocated the important role of GM in human health and disease through several molecular mechanisms. First, the gut microbes have the capacity to increase energy and nutrient extraction from the diet, and it can also alter appetite signaling; second, the human GI flora also offers a physical barrier to protect the host against pathogenic microorganisms, inhibiting their colonization by producing antimicrobial compounds [20][47][48][49]. During metabolism of xenobiotics, the host and its intestinal microbes generates molecules that play crucial part in exchanging information among host cells and its microbial symbionts. Furthermore, there are relevant uremic toxins derived from the intestinal microbial metabolism of proteins, amino acids and other metabolites, including mainly phenols and indoles bounded to proteins and products of metabolism of phenylalanine and tyrosine, such as p-cresol (PC) and p-cresyl sulfate (PCS), and of tryptophan degradation, such as indoxyl sulfate (IS) and indoleacetic acid [50]. Finally, amines and polyamines are also originated by GM metabolism. A major example is choline, a crucial nutrient for lipid metabolism that is metabolized into toxic trimethylamine and further converted to trimethylamine-N-oxide (TMAO) in the liver, which is a promoter of cardiovascular and renal diseases [51][52].
Changed GM composition have been reported in IBDs subjects when compared to controls, though no uniform pattern of alterations has yet been observed [53][54]. The mechanisms underlying the association between GM dysbiosis and evolution of IBDs include the pro-inflammatory and pro-oxidative profile generated in the intestinal lumen and adjacent layers [55][56]. In addition, debates are also going on regarding the crucial role of heat shock proteins (HSPs) in the pathogenesis of IBD, namely due to their participation in several relevant biochemical pathways, such as folding, translocation, and ubiquitinylation of intracellular proteins, as well as due to their capacity to excite innate and adaptive immune response, thus serving as primary autoimmune response targets [57]. The Th17/Treg cells balance, characterized by pro-inflammatory and anti-inflammatory cytokines, which is pivotal for the host’s intestinal homeostasis and induction or inhibition of colonic inflammation, is highly influenced by GM composition. Under inflammatory conditions, including IBD and other GI diseases, antigens derived from a dysbiotic GM activate immune cells (namely Th1 and Th17), causing tissue damage, reduction of mucus layer, and exacerbated penetration of microbes in the intestinal tissues. Consequently, there is an augmented uptake of microbial antigens and toll-like receptor (TLR) ligands that perpetuate the immune responses [8][58].
As above referred, the composition of intestinal microflora is affected by several factors, including lifestyle habits, particularly diet and exercise, which are risk factors for cardiometabolic diseases, such as obesity and T2DM. These conditions have been viewed as the result of an intricate crosstalk between individual genetics, environment influences, including the dietary pattern, as well as the intestinal microflora [59]. Increasing evidences coming from preclinical and clinical studies support the existence of a dysbiotic GM in obesity and T2DM, characterized by lower diversity and resilience [60]. The putative association between dysbiosis and the development of obesity, T2DM and its serious vascular complications has been suggested based on several distinct mechanisms (Figure 2). In brief, GM dysbiosis is a trigger for gut barrier integrity breakdown, with changes on the expression of tight proteins, followed by augmented permeability and consequent translocation from the gut lumen to the bloodstream of bacteria fragments, namely lipopolysaccharide (LPS) and peptidoglycan (PG), and uremic toxins, thus inducing endotoxemia, also referred as a low-grade inflammation state [61][62]. The so-called microbe-associated molecular pattern (MAMP) cause a pro-inflammatory response by binding to toll-like receptors (TLRs), namely TLR4, which evokes a cascade of responses that culminate in pro-inflammatory molecules’ release, which will then affect glucose and insulin metabolism and/or signaling [63]. This is further fueled by advanced glycation end products (AGEs) and other oxidative pathways which are deeply involved in the metabolic impairment in obesity and diabetes development. Concomitantly, several evidences strongly suggest that signals coming from the dysbiotic GM modulate immunometabolism by interfering with epithelial and immune cells, generating an immune-inflammatory milieu that favors the progression of diabetes and its complication [10]. In fact, diabetic gut dysbiosis seems to contribute not only to obesity and diabetes development but also to the progression of some of its main microvascular complications, including diabetic retinopathy (DR) and nephropathy (DN). It is suggested that an increase of circulating bacterial endotoxins, particularly LPS, might play a major role in the low-grade inflammation typical of obesity, diabetes and microvascular complications. The translocation of bacterial components and other microbial-derived products through the impaired intestinal barrier to the systemic circulation can contribute to the pro-inflammatory and pro-oxidative profile found in DN and DR, as well as to the overactivated innate and adaptive immunity [10]. These mechanisms, overall, can be pivotal to the progression of metabolic and vascular complications of diabetes (Figure 2).
The question remains whether dysbiosis is directly linked to metabolic disorders, particularly obesity, T2DM and its vascular complications, or whether the impaired GM composition is an adaptation to alteration of host’s diet and other modulatory factors. There following findings might help to achieve valid answers: (i) microbiota transfer from lean donors into subjects with metabolic disease causes amelioration of insulin sensitivity and (ii) change in human’s diet patterns promote a quick and reversible alteration in the composition of dominant GM members [64][65].
References
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut Microbiota in Health and Disease. Physiol. Rev. 2010, 90, 859–904.
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the Human Intestinal Microbial Flora. Science 2005, 308, 1635–1638.
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207.
- Conlon, M.A.; Bird, A.R. The Impact of Diet and Lifestyle on Gut Microbiota and Human Health. Nutrients 2014, 7, 17–44.
- Krishnan, S.; Alden, N.; Lee, K. Pathways and functions of gut microbiota metabolism impacting host physiology. Curr. Opin. Biotechnol. 2015, 36, 137–145.
- DeGruttola, A.K.; Low, D.; Mizoguchi, A.; Mizoguchi, E. Current Understanding of Dysbiosis in Disease in Human and Animal Models. Inflamm. Bowel Dis. 2016, 22, 1137–1150.
- Zhang, M.; Sun, K.; Wu, Y.; Yang, Y.; Tso, P.; Wu, Z. Interactions between Intestinal Microbiota and Host Immune Response in Inflammatory Bowel Disease. Front. Immunol. 2017, 8, 942.
- Zuo, T.; Ng, S.C. The Gut Microbiota in the Pathogenesis and Therapeutics of Inflammatory Bowel Disease. Front. Microbiol. 2018, 9, 2247.
- Brial, F.; Le Lay, A.; Dumas, M.-E.; Gauguier, D. Implication of gut microbiota metabolites in cardiovascular and metabolic diseases. Cell. Mol. Life Sci. 2018, 75, 3977–3990.
- Fernandes, R.; Viana, S.D.; Nunes, S.; Reis, F. Diabetic gut microbiota dysbiosis as an inflammaging and immunosenescence condition that fosters progression of retinopathy and nephropathy. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1865, 1876–1897.
- Pascale, A.; Marchesi, N.; Marelli, C.; Coppola, A.; Luzi, L.; Govoni, S.; Giustina, A.; Gazzaruso, C. Microbiota and metabolic diseases. Endocrine 2018, 61, 357–371.
- Crasci, L.; Lauro, M.R.; Puglisi, G.; Panico, A. Natural antioxidant polyphenols on inflammation management: Anti-glycation activity vs. metalloproteinases inhibition. Crit. Rev. Food Sci. Nutr. 2018, 58, 893–904.
- Kinger, M.; Kumar, S.; Kumar, V. Some Important Dietary Polyphenolic Compounds: An Anti-inflammatory and Immunoregulatory Perspective. Mini Rev. Med. Chem. 2018, 18, 1270–1282.
- Sharma, U.K.; Sharma, A.K.; Pandey, A.K. Medicinal attributes of major phenylpropanoids present in cinnamon. BMC Complement. Altern. Med. 2016, 16, 156.
- Singh, A.K.; Bishayee, A.; Pandey, A.K. Targeting Histone Deacetylases with Natural and Synthetic Agents: An Emerging Anticancer Strategy. Nutrients 2018, 10, 731.
- Landete, J.M. Updated Knowledge about Polyphenols: Functions, Bioavailability, Metabolism, and Health. Crit. Rev. Food Sci. Nutr. 2012, 52, 936–948.
- Duda-Chodak, A.; Tarko, T.; Satora, P.; Sroka, P. Interaction of dietary compounds, especially polyphenols, with the intestinal microbiota: A review. Eur. J. Nutr. 2015, 54, 325–341.
- Kawabata, K.; Yoshioka, Y.; Terao, J. Role of Intestinal Microbiota in the Bioavailability and Physiological Functions of Dietary Polyphenols. Molecules 2019, 24, 370.
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836.
- Gill, S.R.; Pop, M.; DeBoy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic Analysis of the Human Distal Gut Microbiome. Science 2006, 312, 1355–1359.
- Ursell, L.K.; Haiser, H.J.; Van Treuren, W.; Garg, N.; Reddivari, L.; Vanamala, J.; Dorrestein, P.C.; Turnbaugh, P.J.; Knight, R. The Intestinal Metabolome: An Intersection Between Microbiota and Host. Gastroenterology 2014, 146, 1470–1476.
- Li, H.; Limenitakis, J.P.; Fuhrer, T.; Geuking, M.B.; Lawson, M.A.; Wyss, M.; Brugiroux, S.; Keller, I.; MacPherson, J.A.; Rupp, S.; et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat. Commun. 2015, 6, 8292.
- DiGiulio, D.B.; Romero, R.; Amogan, H.P.; Kusanovic, J.P.; Bik, E.M.; Gotsch, F.; Kim, C.J.; Erez, O.; Edwin, S.; Relman, D.A. Microbial Prevalence, Diversity and Abundance in Amniotic Fluid During Preterm Labor: A Molecular and Culture-Based Investigation. PLoS ONE 2008, 3, e3056.
- Koleva, P.T.; Kim, J.; Scott, J.A.; Kozyrskyj, A.L. Microbial programming of health and disease starts during fetal life. Birth Defects Res. Part C Embryo Today Rev. 2015, 105, 265–277.
- Satokari, R.; Grönroos, T.; Laitinen, K.; Salminen, S.; Isolauri, E. BifidobacteriumandLactobacillusDNA in the human placenta. Lett. Appl. Microbiol. 2009, 48, 8–12.
- Marchesi, J.R.; Adams, D.H.; Fava, F.; Hermes, G.D.; Hirschfield, G.M.; Hold, G.; Quraishi, M.N.; Kinross, J.; Smidt, H.; Tuohy, K.M.; et al. The gut microbiota and host health: A new clinical frontier. Gut 2016, 65, 330–339.
- Tanaka, M.; Nakayama, J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol. Int. 2017, 66, 515–522.
- Bull, M.J.; Plummer, N.T. Part 1: The Human Gut Microbiome in Health and Disease. Integr. Med. A Clin. J. 2014, 13, 17.
- Wu, H.-J.; Wu, E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 2012, 3, 4–14.
- Kim, D.; Zeng, M.Y.; Núñez, G. The interplay between host immune cells and gut microbiota in chronic inflammatory diseases. Exp. Mol. Med. 2017, 49, e339.
- Belkaid, Y.; Hand, T.W. Role of the Microbiota in Immunity and Inflammation. Cell 2014, 157, 121–141.
- Koenig, J.E.; Spor, A.; Scalfone, N.; Fricker, A.D.; Stombaugh, J.; Knight, R.; Angenent, L.T.; Ley, R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108, 4578–4585.
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227.
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696.
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230.
- An, R.; Wilms, E.; Masclee, A.A.M.; Smidt, H.; Zoetendal, E.G.; Jonkers, D. Age-dependent changes in GI physiology and microbiota: Time to reconsider? Gut 2018, 67, 2213–2222.
- Mariat, D.; Firmesse, O.; Levenez, F.; Guimarăes, V.; Sokol, H.; Doré, J.; Corthier, G.; Furet, J.-P. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009, 9, 123.
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Harris, H.M.B.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184.
- Harmsen, H.J.M.; Wildeboer–Veloo, A.C.M.; Raangs, G.C.; Wagendorp, A.A.; Klijn, N.; Bindels, J.G.; Welling, G.W. Analysis of Intestinal Flora Development in Breast-Fed and Formula-Fed Infants by Using Molecular Identification and Detection Methods. J. Pediatr. Gastroenterol. Nutr. 2000, 30, 61–67.
- Tremaroli, V.; Bäckhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249.
- Bian, X.; Chi, L.; Gao, B.; Tu, P.; Ru, H.; Lu, K. Gut Microbiome Response to Sucralose and Its Potential Role in Inducing Liver Inflammation in Mice. Front. Physiol. 2017, 8, 487.
- Chassaing, B.; Koren, O.; Goodrich, J.K.; Poole, A.C.; Srinivasan, S.; Ley, R.E.; Gewirtz, A.T. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 2015, 519, 92–96.
- Wu, G.D.; Compher, C.; Chen, E.Z.; Smith, S.A.; Shah, R.D.; Bittinger, K.; Chehoud, C.; Albenberg, L.G.; Nessel, L.; Gilroy, E.; et al. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut 2016, 65, 63–72.
- Looft, T.; Allen, H.K. Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes 2012, 3, 463–467.
- Willing, B.P.; Russell, S.L.; Finlay, B.B. Shifting the balance: Antibiotic effects on host–microbiota mutualism. Nat. Rev. Genet. 2011, 9, 233–243.
- Modi, S.R.; Collins, J.J.; Relman, D.A. Antibiotics and the gut microbiota. J. Clin. Investig. 2014, 124, 4212–4218.
- Cash, H.L.; Whitham, C.V.; Behrendt, C.L.; Hooper, L.V. Symbiotic Bacteria Direct Expression of an Intestinal Bactericidal Lectin. Science 2006, 313, 1126–1130.
- Hooper, L.V.; Stappenbeck, T.S.; Hong, C.V.; Gordon, J.I. Angiogenins: A new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 2003, 4, 269–273.
- Schauber, J.; Svanholm, C.; Termén, S.; Iffland, K.; Menzel, T.; Scheppach, W.; Melcher, R.; Agerberth, B.; Lührs, H.; Gudmundsson, G.H. Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: Relevance of signalling pathways. Gut 2003, 52, 735–741.
- Evenepoel, P.; Meijers, B.K.; Bammens, B.R.; Verbeke, K. Uremic toxins originating from colonic microbial metabolism. Kidney Int. 2009, 76, S12–S19.
- Bennett, B.J.; Vallim, T.Q.D.A.; Wang, Z.; Shih, D.M.; Meng, Y.; Gregory, J.; Allayee, H.; Lee, R.; Graham, M.; Crooke, R.; et al. Trimethylamine-N-Oxide, a Metabolite Associated with Atherosclerosis, Exhibits Complex Genetic and Dietary Regulation. Cell Metab. 2013, 17, 49–60.
- Mutsaers, H.A.M.; Stribos, E.G.D.; Glorieux, G.; Vanholder, R.; Olinga, P. Chronic Kidney Disease and Fibrosis: The Role of Uremic Retention Solutes. Front. Med. 2015, 2, 60.
- Krogius-Kurikka, L.; Lyra, A.; Malinen, E.; Aarnikunnas, J.; Tuimala, J.; Paulin, L.; Mäkivuokko, H.; Kajander, K.; Palva, A. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009, 9, 95.
- Salonen, A.; De Vos, W.M.; Palva, A. Gastrointestinal microbiota in irritable bowel syndrome: Present state and perspectives. Microbiology 2010, 156, 3205–3215.
- Ananthakrishnan, A.N. Epidemiology and risk factors for IBD. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 205–217.
- Wędrychowicz, A.; Zając, A.; Tomasik, P. Advances in nutritional therapy in inflammatory bowel diseases: Review. World J. Gastroenterol. 2016, 22, 1045–1066.
- Bellavia, M.; Tomasello, G.; Romeo, M.; Damiani, P.; Monte, A.I.L.; Lozio, L.; Campanella, C.; Gammazza, A.M.; Rappa, F.; Zummo, G.; et al. Gut microbiota imbalance and chaperoning system malfunction are central to ulcerative colitis pathogenesis and can be counteracted with specifically designed probiotics: A working hypothesis. Med. Microbiol. Immunol. 2013, 202, 393–406.
- SAME AS 8 Khan, I.; Ullah, N.; Zha, L.; Bai, Y.; Khan, A.; Zhao, T.; Che, T.; Zhang, C. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens 2019, 8, 126.
- Franks, P.W.; McCarthy, M.I. Exposing the exposures responsible for type 2 diabetes and obesity. Science 2016, 354, 69–73.
- Lazar, V.; Ditu, L.M.; Pircalabioru, G.G.; Picu, A.; Petcu, L.; Cucu, N.; Chifiriuc, M.C. Gut Microbiota, Host Organism, and Diet Trialogue in Diabetes and Obesity. Front. Nutr. 2019, 6, 21.
- Burcelin, R. Gut microbiota and immune crosstalk in metabolic disease. Mol. Metab. 2016, 5, 771–781.
- Gomes, J.M.G.; Costa, J.A.; Alfenas, R.C.G. Metabolic endotoxemia and diabetes mellitus: A systematic review. Metabolism 2017, 68, 133–144.
- Blandino, G.; Inturri, R.; Lazzara, F.; Di Rosa, M.; Malaguarnera, L. Impact of gut microbiota on diabetes mellitus. Diabetes Metab. 2016, 42, 303–315.
- Vrieze, A.; Van Nood, E.; Holleman, F.; Salojärvi, J.; Kootte, R.S.; Bartelsman, J.F.; Dallinga–Thie, G.M.; Ackermans, M.T.; Serlie, M.J.; Oozeer, R.; et al. Transfer of Intestinal Microbiota from Lean Donors Increases Insulin Sensitivity in Individuals with Metabolic Syndrome. Gastroenterology 2012, 143, 913–916.
- Walker, A.W.; Ince, J.; Duncan, S.H.; Webster, L.M.; Holtrop, G.; Ze, X.; Brown, D.; Stares, M.D.; Scott, P.; Bergerat, A.; et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011, 5, 220.
More
Information
Subjects:
Nutrition & Dietetics
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
991
Entry Collection:
Gastrointestinal Disease
Revisions:
2 times
(View History)
Update Date:
26 Aug 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




