Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jose Antonio Curiel | + 2095 word(s) | 2095 | 2021-08-12 08:49:04 | | | |
| 2 | Rita Xu | + 2057 word(s) | 2057 | 2021-08-12 09:35:20 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Curiel, J.A. Bifidobacterial α-L-Fucosidases. Encyclopedia. Available online: https://encyclopedia.pub/entry/13090 (accessed on 07 February 2026).
Curiel JA. Bifidobacterial α-L-Fucosidases. Encyclopedia. Available at: https://encyclopedia.pub/entry/13090. Accessed February 07, 2026.
Curiel, Jose Antonio. "Bifidobacterial α-L-Fucosidases" Encyclopedia, https://encyclopedia.pub/entry/13090 (accessed February 07, 2026).
Curiel, J.A. (2021, August 12). Bifidobacterial α-L-Fucosidases. In Encyclopedia. https://encyclopedia.pub/entry/13090
Curiel, Jose Antonio. "Bifidobacterial α-L-Fucosidases." Encyclopedia. Web. 12 August, 2021.
Copy Citation
Fucosylated carbohydrates and glycoproteins from human breast milk are essential for the development of the gut microbiota in early life because they are selectively metabolized by bifidobacteria.
bifidobacteria
fucosidases
glycosyl hydrolases
1. Introduction
The impact of human milk glycobiome on the gut microbiota of infants is well established [1]. While a great part of the components of breast milk provide nutrients to the infant, human milk oligosaccharides (HMOs) and human milk glycoproteins (HMGs) selectively favor the colonization and growth of bifidobacteria in the infant intestine, contributing to the development of the gut microbiota [1][2]. In this regard, Bifidobacterium species are considered key actors in the multifaceted process of gut development and maturation of the immune system [3]. In fact, during the first months of birth, the loss of bifidobacteria or the gain of other bacteria can significantly alter the progression of the healthy microbial community with negative consequences for the infant, including a predisposition to autoimmune and/or metabolic diseases such as allergies and childhood obesity [4][5]. Concerning to that, fucosylated HMOs (FHMOs) and fucosylated HMGs (FHMGs) constitute a great part of the glycobiome of the breast milk [6] (Figure 1) and have been proposed to be essential in the development of the microbiota [7].
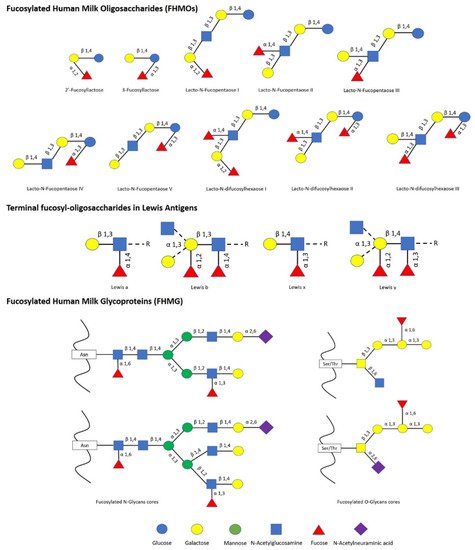
FHMOs constitute the largest fraction of human milk oligosaccharides, and although they show a small number of different conformations, they can make up to 70% of the total in an individual mother’s milk [6]. The fucosylated trisaccharide 2′-fucosyllactose is the most abundant FHMO, representing from 12 to 45% of the total HMO content in breastmilk, while 3-fucosyllactose is less abundant, from 0.5% to 3% [8]. On the other hand, there are several FMHGs investigated, and contrary to FHMOs, they appear at lower concentration but show a higher number of different forms, including lactoferrin (17%), immunoglobulins IgG (<1%), IgM (<1%), and secretory IgA (11%) [9][10][11]. Both FHMOs and FHMGs stand out for their ability to stimulate the growth of bifidobacteria [7][12], whose metabolism transforms fucosylated oligosaccharides into short-chain fatty acids (SCFAs) such as acetate, formate, lactate, and pyruvate [13], which in turn stimulate the immune system by inducing the differentiation of T-regulatory cells via inhibition of histone deacetylase [14].
The great influence of fucosylated compounds present in breast milk on bifidobacteria is due to their ability to metabolize them, being α-L-fucosidases (henceforth, fucosidases) indispensable tools that allow shaping the gut microbiome in the first months of life.
According to CAZy database, hitherto, more than 10,000 sequences have been identified in silico as α-L-fucosidases, belonging to a wide variety of organisms from archaea to fungi and plants. However, the vast majority of fucosidase sequences have been described in bacteria and belong to more than 2000 bacteria species (www.cazy.org). This database classifies fucosidases into three families (GH29, GH95, and GH151) according to their catalytic structures. GH29 fucosidases act through a retaining mechanism and have a broader substrate specificity, including hydrolysis of Fuc-α1,3/4/6 linkages [15]. Moreover, family GH29 fucosidases have been subclassified into two subfamilies. The subfamily A contains α-fucosidases with relatively relaxed substrate specificities, able to hydrolyze p-nitrophenyl-α-L-fucopyranoside (pNP-fucose), while the members of subfamily B are specific to α1,3/4-glycosidic linkages and are practically unable to hydrolyze pNP-fucose [16]. Although GH29 fucosidases also could exhibit hydrolysis of Fuc-α1,2 linkages, that activity is mainly attributed to GH95 family, which catalyzes the hydrolysis of fucose linkages through an inverting mechanism, resulting in the inversion of the anomeric configuration [17][18]. Finally, GH151 family has poor activity on fucosylated substrates; this is the reason why it is currently questioned as to whether they are genuine fucosidases [19][20][21].
Even though species of the Bifidobacterium genus dominate the infant gut microbiota in early life, and given the importance of their metabolism of fucosylated conjugates, there are only a few bifidobacterial species studied extensively at both cellular and genomics level for their ability to utilize fucosylated carbohydrates, including B. bifidum and Bifidobacterium longum subsp. infantis [22][23]. However, different strain-dependent metabolic abilities have been unraveled for the use of fucosylated conjugates and are likely determined by their fucosidases’ diversity [24]. Indeed, agreeing with the evolution and phylogenetics of fucosidases previously studied in metazoan fucosidases [25], bifidobacterial fucosidase sequences listed in CAZy reveal substantial in silico differences regarding to their conserved domains, even those ones clustered in the same GH, revealing different adaptation/specialization ranges as well as their origin.
2. Bifidobacterial GH29 Fucosidases
Based on in silico studies concerning conserved domains released by NCBI Conserved Domains Database (CDD), bifidobacterial GH29 fucosidases could be classified into four different phylogenetic groups. That differentiation was also confirmed through sequence homology PCA and cluster analyses (Figure 2).
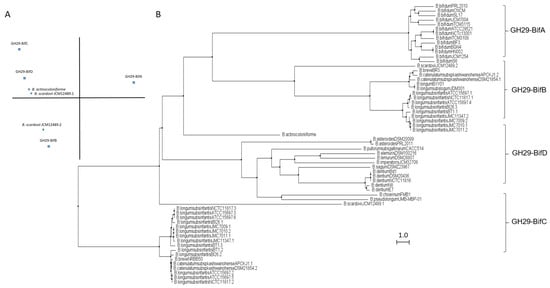
Figure 2. Phylogenetic analysis of bifidobacterial GH29 fucosidases. PCA (A) and cladogram tree (B) distributions of bifidobacterial GH29 fucosidase sequences listed in CAZy, released from Jalview 2.11.1.4 software using the neighbor-joining method.
The enzymes included in the proposed GH29-BifA, only found in B. bifidum strains, are characterized as large membrane-bound fucosidases (AfuC super family domain; NCBI CDD accession number cl34656) and exhibit an accessory F5/F8 type C domain family (NCBI CDD accession number cl23730), probably involved in recognizing galactose or N-acetyllactosamine [26]. Interestingly, while InterPro database (EMBL-EBI) recognized the F5/F8 type C domain (IPR000421), it interpreted the AfuC domain as Glyco_Hydro_29 domain (IPR000933), probably due to the degree of updating of both databases. In addition, Ashida et al., 2009 identified a second putative sugar-binding domain in GH29 fucosidase AfcB from B. bifidum JCM1254, domain that is frequently found in membrane-bound or cell-wall-associated proteins and denominated FIVAR [27]. Those results were here confirmed by SOSUI and HMMTOP databases, which allowed the identification of two putative transmembrane helices in GH29-BifA fucosidases. Therefore, it has been suggested that both accessory F5/F8 type C and FIVAR domains allow the extracellular character of GH29 fucosidases in B. bifidum and could enhance affinity toward fucosyl conjugates [27]. Moreover, in all the N-terminal regions of GH29-BifA fucosidases, hydrophobic sequences predicted by SignalP-5.0 to be putative signal peptide with potential cleavage sites were observed.
Concerning the AfuC/Glyco_hydro_29 domain, the only representative GH29 fucosidase of GH29-BifA purified and characterized, which is AfcB from B. bifidum ATCC 1254, is able to hydrolyze 3-fucosyllactose, Lewis blood group substances (a, b, x, and y types), and lacto-N-fucopentaose II and III. However, the enzyme did not act on glycoconjugates containing α1,2-fucosyl residue or on synthetic pNP-fucose [27].
Supporting the in silico characterization of GH29-BifA fucosidases, several studies confirm the ability of B. bifidum to extracellularly hydrolyze FHMOs [28]. However, B. bifidum appears to prefer the utilization of lactose when growing on FHMO, probably releasing fucose to the environment [28]. This incapacity to consume fucose may be due to the lack of specific transporters. Nevertheless, the extracellular fucosidase activity of B. bifidum could facilitate the establishment of the bifidobacteria community, allowing them to consume the released fucose residues [29].
In contrast to GH29-BifA, the rest of the GH29 fucosidases from bifidobacteria do not have either putative signal peptides or transmembrane helices and consequently their mode of action can be considered intracellular. Indeed, GH29-BifB fucosidases are characterized by exhibiting an AfuC super family/Glyco_Hydro_29 domain (NCBI CDD accession number cl34656/IPR000933) such as GH29-BifA fucosidases but lacking F5/8 type C and FIVAR domains. Due to the presence of the same fucosidase domain in both groups of fucosidases (GH29-BifA and GH29-BifB), similar metabolic capacities could be affirmed. In fact, the only characterized bifidobacterial GH29-BifB fucosidase (Blon_2336 from Bifidobacterium longum subsp. infantis ATCC 15697) revealed similar activity to AfcB from B. bifidum ATCC 1254 (GH29-BifA) against Fuc-α1,3 glucosidic, Fuc-α1,3GlcNAc, and Fuc-α1,4GlcNAc linkages [21]. These GH29-BifB fucosidases appear to be distributed along strains of different species, contrary to GH29-BifA fucosidases, and frequently, strains that exhibit GH29-BifB fucosidases also show GH29-BifC fucosidases, which are duplicated in some of the sequenced strains. Actually, the duplication of GH29 fucosidases has been reported previously and plays an important role in fucosidases evolution [30].
GH29-BifC fucosidases are characterized by showing conserved α-Amylase catalytic domain family (NCBI CDD accession number cl38930). It must be taken into account that this superfamily is present in a large number of GHs able to hydrolyze α1,4/6 glycosidic bonds, although in turn they have specific domains unlike the GH29-BifC fucosidases of bifidobacteria [31]. However, since GH29-BifC fucosidases can catalyze the transformation of fucosidic α1-2Gal/3GlcNAc linkages in LNFP I and III, respectively, and mainly Fuc-α1,6 GlcNAc linkages [32], activity non described in the above fucosidase groups, it is difficult to ensure that its catalytic family proposed is α-Amylase catalytic (NCBI CDD) or Glyco_Hydro_29 (InterPro). In this sense, InterPro database (EMBL-EBI) indicated the presence in GH29-BifC fucosidases of a second catalytic family denominated FUC_metazoa_typ (IPR016286) that is close to eukaryotic fucosidases. Probably the presence of this domain is key for these fucosidases to be considered as the most unspecific and versatile fucosidases of bifidobacteria since a wide range of substrates has been reported for two different GH29-BifC fucosidases from B. longum subsp. infantis ATCC 15697 [21][27].
Both GH29-BifB/C fucosidases described in B. longum subsp. infantis strains are likely found in the cytosol. Therefore, efficient transport of oligosaccharides is needed, unlike B. bifidum [13][21]. In this context, genomic studies carried out on B. longum subsp. infantis ATCC 15697 have unraveled several putative fucose permeases that may facilitate environmental scavenging when soluble fucose is encountered.
In order to elucidate the roles and fitness of the bifidobacterial community to shape the gut microbiome and taking into account the relevance of fucosidases in this regard, their features mentioned above should be updated and expanded to avoid ambiguities in the catalytic domains and relate them to their metabolic properties. Certainly, the rest of the enzymes from different bifidobacterial species need to be characterized in order to reliably distinguish the properties of each group of fucosidases for determining the interaction and mode of actions of bifidobacteria during gut colonization. In this sense, the role of GH29-BifD of fucosidases remains unknown despite having been sequenced and identified in certain Bifidobacterium species. Unlike to GH29-BifC, GH29-BifD fucosidases exhibit specific α-L-fucosidase main domain (NCBI CDD accession number cl38930). Surprisingly, their accession number is matching with superfamily AmyAc family of group II, suggesting a better accurate and updated in silico annotation. However, InterPro database (EMBL_EBI) indicates both catalytic domain Glyco_Hydro_29 and FUC_metazoa_typ (InterPro IPR000933 and IPR016286, respectively). Nevertheless, physicochemical properties, substrate specificity confirmation, and their correlation with catalytic domains are still pending to be characterized.
3. Bifidobacterial GH95 Fucosidases
Similar to GH29 bifidobacterial fucosidases and according to architecture domains, bifidobacterial GH95 fucosidases collected on CAZy could also be subclassified into two main groups (Figure 3).
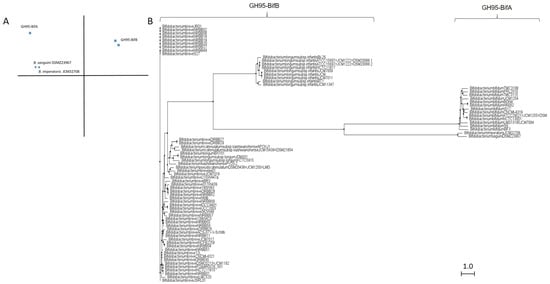
Figure 3. Phylogenetic analysis of bifidobacterial GH95 fucosidases. PCA (A) and cladogram tree (B) distributions of bifidobacterial GH95 fucosidase sequences listed in CAZy, released from Jalview 2.11.1.4 software using the neighbor-joining method.
The extracellular character observed in GH29-BifA fucosidases from B. bifidum strains is also reflected in their GH95 fucosidases, which are characterized by a putative signal peptide and two predicted transmembrane helices. Among GH95 fucosidases, those features are only found in the proposed GH95-BifA fucosidases from B. bifidum with the exception of Bifidobacterium saguini DSMZ 23967 fucosidase (Genbank QTB91571.1), which exhibited two putative transmembrane helices.
The proposed GH95-BifA was characterized according to the NCBI CDD database by exhibiting Glycosyl hydrolase 65 N-terminal (accession number cl22392) as main catalytic domain, while InterPro database analysis (EMBL-EBI) revealed a Glycosyl hydrolase 95 N-terminal (IPR027414). The observed ambiguous prediction on the catalytic architecture could be due to the lack of updating and mismatch annotations. Nevertheless, a common evolutionary origin for GH65 and GH95 families, among others, with conservation of their putative catalytic amino acid residues, was noticed and likely influenced the in silico results [18]. Nevertheless, and contrary to GH65 family, the only GH95-BifA representative fucosidase recombinantly produced and characterized (AfcA from B. bifidum JCM1254) showed great activity against Fuc-α1,2 Gal linkages, mainly hydrolyzing 2′-Fucosyllactose and lacto-N-fucopentaose I [17][33].
On the other hand, while NCBI CDD database detected two YjdB overlapping domains (accession number cl35007), whose functions are still uncharacterized but in turn contain Ig-like domain, InterPro database noticed Ig-like_Bact and Bacterial Ig-like group 2 (BIG2) domains instead (accession number IPR022038 and IPR003343, respectively). Despite this coincidence, only the position of one domain practically matches in both databases (YjdB and BIG2). In addition, InterPro identifies Ig-like_Bact near to N-terminal unlike NCBI CDD, and probably GH95-BifA sequences could exhibit up to three accessory domains.
References
- Zivkovic, A.M.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc. Natl. Acad. Sci. USA 2011, 108, 4653–4658.
- Milani, C.; Mancabelli, L.; Lugli, G.A.; Duranti, S.; Turroni, F.; Ferrario, C.; Mangifesta, M.; Viappiani, A.; Ferreti, P.; Corfer, V.; et al. Exploring vertical transmission of bifidobacteria from mother to child. Appl. Environ. Microbiol. 2015, 81, 7078–7087.
- Turroni, F.; Peano, C.; Pass, D.A.; Foroni, E.; Severgnini, M.; Claesson, M.J.; Kerr, C.; Hourihane, J.; Murray, D.; Fuligni, F.; et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE 2012, 7, e36957.
- O’Neill, I.; Schofield, Z.; Hall, L.J. Exploring the role of the microbiota member Bifidobacterium in modulating immune-linked diseases. Emerg. Top. Life Sci. 2017, 1, 333–349.
- Wampach, L.; Heintz-Buschart, A.; Fritz, J.V.; Ramiro-Garcia, J.; Habier, J.; Herold, M.; Nayaranasamy, S.; Kaysen, A.; Hogan, A.H.; Bindl, L.; et al. Birth mode determines earliest strainconferred gut microbiome functions and immunostimulatory potential. Nat. Commun. 2018, 9, 1–14.
- Wu, S.; Tao, N.; German, J.B.; Grimm, R.; Lebrilla, C.B. Development of an annotated library of neutral human milk oligosaccharides. J. Proteome Res. 2010, 9, 4138–4151.
- Bai, Y.; Tao, J.; Zhou, J.; Fan, Q.; Liu, M.; Hu, Y.; Xu, Y.; Zhang, L.; Yuan, J.; Li, W.; et al. Fucosylated human milk oligosaccharides and N-glycans in the milk of Chinese mothers regulate the gut microbiome of their breast-fed infants during different lactations stages. MSystems 2018, 3, e00206-18.
- McGuire, M.K.; Meehan, C.L.; McGuire, M.A.; Williams, J.E.; Foster, J.; Sellen, D.W.; Prentice, A.M. What’s normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am. J. Clin. Nutr. 2017, 105, 1086–1100.
- Huang, J.; Guerrero, A.; Parker, E.; Strum, J.S.; Smilowitz, J.T.; German, J.B.; Lebrilla, C.B. Site-specific glycosylation of secretory immunoglobulin A from human colostrum. J. Proteome Res. 2015, 14, 1335–1349.
- Li, W.; Yu, R.; Ma, B.; Yang, Y.; Jiao, X.; Liu, Y.; Cao, H.; Dong, W.; Liu, L.; Ma, K.; et al. Core fucosylation of IgG B cell receptor is required for antigen recognition and antibody production. J. Immunol. 2015, 194, 2596–2606.
- Peterson, R.; Cheah, W.Y.; Grinyer, J.; Packer, N. Glycoconjugates in human milk: Protecting infants from disease. Glycobiology 2013, 23, 1425–1438.
- Korpela, K.; Salonen, A.; Hickman, B.; Kunz, C.; Sprenger, N.; Kukkonen, K.; Savilahti, E.; Kuitunen, M.; de Vos, W.M. Fucosylated oligosaccharides in mother’s milk alleviate the effects of caesarean birth on infant gut microbiota. Sci. Rep. 2018, 8, 1–7.
- Zabel, B.E.; Gerdes, S.; Evans, K.C.; Nedveck, D.; Singles, S.K.; Volk, B.; Budinoff, C. Strain-specific strategies of 2′-fucosyllactose, 3-fucosyllactose, and difucosyllactose assimilation by Bifidobacterium longum subsp. infantis Bi-26 and ATCC 15697. Sci. Rep. 2020, 10, 1–18.
- Park, J.; Kim, M.; Kang, S.G.; Jannasch, A.H.; Cooper, B.; Patterson, J.; Kim, C.H. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR–S6K pathway. Mucosal Immunol. 2015, 8, 80–93.
- Grootaert, H.; Van Landuyt, L.; Hulpiau, P.; Callewaert, N. Functional exploration of the GH29 fucosidase family. Glycobiology 2020, 30, 735–745.
- Shaikh, F.A.; Van Bueren, A.L.; Davies, G.J.; Withers, S.G. Identifying the catalytic acid/base in GH29 α-L-Fucosidase subfamilies. Biochemistry 2013, 52, 5857–5864.
- Katayama, T.; Sakuma, A.; Kimura, T.; Makimura, Y.; Hiratake, J.; Sakata, K.; Yamanoi, T.; Kumagai, H.; Yamamoto, K. Molecular cloning and characterization of Bifidobacterium bifidum 1, 2-α-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95). J. Bacteriol. 2004, 186, 4885–4893.
- Nagae, M.; Tsuchiya, A.; Katayama, T.; Yamamoto, K.; Wakatsuki, S.; Kato, R. Structural basis of the catalytic reaction mechanism of novel 1, 2-α-L-fucosidase from Bifidobacterium bifidum. J. Biol. Chem. 2007, 282, 18497–18509.
- Lombard, V.; Ramulu, H.G.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, 490–495.
- Lezyk, M.; Jers, C.; Kjaerulff, L.; Gotfredsen, C.H.; Mikkelsen, M.D.; Mikkelsen, J.D. Novel α-L-fucosidases from a soil metagenome for production of fucosylated human milk oligosaccharides. PLoS ONE 2016, 11, e0147438.
- Sela, D.A.; Garrido, D.; Lerno, L.; Wu, S.; Tan, K.; Eom, H.J.; Joachimiak, A.; Lebrilla, C.B.; Mills, D.A. Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl. Environ. Microbiol. 2012, 78, 795–803.
- Angeloni, S.; Ridet, J.L.; Kusy, N.; Gao, H.; Crevoisier, F.; Guinchard, S.; Kochhar, S.; Sigrist, H.; Sprenger, N. Glycoprofiling with micro-arrays of glycoconjugates and lectins. Glycobiology 2005, 15, 31–41.
- Bergström, A.; Skov, T.H.; Bahl, M.I.; Roager, H.M.; Christensen, L.B.; Ejlerskov, K.T.; Molgaard, C.; Michaelsen, K.F.; Licht, T.R. Establishment of intestinal microbiota during early life: A longitudinal, explorative study of a large cohort of Danish infants. Appl. Environ. Microbiol. 2014, 80, 2889–2900.
- Turroni, F.; Bottacini, F.; Foroni, E.; Mulder, I.; Kim, J.H.; Zomer, A.; Sánchez, B.; Bidossi, A.; Ferrarini, A.; Giubellini, V.; et al. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. USA 2010, 107, 19514–19519.
- Intra, J.; Perotti, M.E.; Pavesi, G.; Horner, D. Comparative and phylogenetic analysis of α-l-fucosidase genes. Gene 2007, 392, 34–46.
- Abbott, D.W.; Eirin-Lopez, J.M.; Boraston, A.B. Insight into ligand diversity and novel biological roles for family 32 carbohydrate-binding modules. Mol. Biol. Evol. 2008, 25, 155–167.
- Ashida, H.; Miyake, A.; Kiyohara, M.; Wada, J.; Yoshida, E.; Kumagai, H.; Katayama, T.; Yamamoto, K. Two distinct α-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology 2009, 19, 1010–1017.
- Garrido, D.; Ruiz-Moyano, S.; Lemay, D.G.; Sela, D.A.; German, J.B.; Mills, D.A. Comparative transcriptomics reveals key differences in the response to milk oligosaccharides of infant gut-associated bifidobacteria. Sci. Rep. 2015, 5, 1–18.
- Asakuma, S.; Hatakeyama, E.; Urashima, T.; Yoshida, E.; Katayama, T.; Yamamoto, K.; Kumagai, H.; Ashida, H.; Hirose, J.; Kitaoka, M. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J. Biol. Chem. 2011, 286, 34583–34592.
- You, J.; Lin, S.; Jiang, T. Origins and evolution of the α-L-fucosidases: From bacteria to metazoans. Front. Microbiol. 2019, 10, 1756.
- Janeček, Š.; Svensson, B.; MacGregor, E.A. α-Amylase: An enzyme specificity found in various families of glycoside hydrolases. Cell. Mol. Life Sci. 2014, 71, 1149–1170.
- Ashida, H.; Fujimoto, T.; Kurihara, S.; Nakamura, M.; Komeno, M.; Huang, Y.; Katayama, T.; Kinoshita, T.; Takegawa, K. 1,6-α-L-Fucosidases from Bifidobacterium longum subsp. infantis ATCC 15697 involved in the degradation of core-fucosylated N-glycan. J. Appl. Glycosci. 2020, 67, 23–29.
- Gotoh, A.; Katoh, T.; Sakanaka, M.; Ling, Y.; Yamada, C.; Asakuma, S.; Urashima, T.; Tomabechi, Y.; Katayama-Ikegami, A.; Kurihara, S.; et al. Sharing of human milk oligosaccharides degradants within Bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum. Sci. Rep. 2018, 8, 1–14.
More
Information
Subjects:
Biophysics
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
898
Revisions:
2 times
(View History)
Update Date:
12 Aug 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




