Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Alessandro Poli | + 2585 word(s) | 2585 | 2021-06-01 08:03:18 | | | |
| 2 | Alessandro Poli | Meta information modification | 2585 | 2021-08-11 11:58:04 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Poli, A. Phosphatidylinositol 5 Phosphate. Encyclopedia. Available online: https://encyclopedia.pub/entry/12746 (accessed on 08 February 2026).
Poli A. Phosphatidylinositol 5 Phosphate. Encyclopedia. Available at: https://encyclopedia.pub/entry/12746. Accessed February 08, 2026.
Poli, Alessandro. "Phosphatidylinositol 5 Phosphate" Encyclopedia, https://encyclopedia.pub/entry/12746 (accessed February 08, 2026).
Poli, A. (2021, August 04). Phosphatidylinositol 5 Phosphate. In Encyclopedia. https://encyclopedia.pub/entry/12746
Poli, Alessandro. "Phosphatidylinositol 5 Phosphate." Encyclopedia. Web. 04 August, 2021.
Copy Citation
Phosphatidylinositol (PI)-related signaling plays a pivotal role in many cellular aspects, including survival, cell proliferation, differentiation, DNA damage, and trafficking. PI is the core of a network of proteins represented by kinases, phosphatases, and lipases which are able to add, remove or hydrolyze PI, leading to different phosphoinositide products. Among the seven known phosphoinositides, phosphatidylinositol 5 phosphate (PI5P) was the last to be discovered. PI5P presence in cells is very low compared to other PIs, but is has been reported to control many cellular outcomes, including cell proliferation, gene expression and chromatin remodeling.
PI5P
PIKFyve
myotubularin
PI5P4K/PIP4K
phosphatases
nucleus
1. Introduction
1.1. Phosphatidylinositol Signaling
Multiple cellular functions, including survival, proliferation, differentiation, DNA damage response, and gene transcription can be modulated by a specific network of kinases, phosphatases, and lipases able to modulate the lipid second messenger phosphatidylinositol [1][2][3][4][5]. Phosphatidylinositol (PI) is composed of two different modules: a hydrophilic inositol head group bound through a phosphodiester bond to a glycerol and two fatty acids tails that represent the hydrophobic part of the molecule. The fatty acids tails are prevalently represented by stearic and arachidonic acids but other acyl chains are known to be present [6]. Modifications of the inositol ring due to addition or removal of phosphate groups, together with hydrolysis of the phosphodiester bond by phospholipases C, are the most common changes leading to the production of second messengers involved in many cellular aspects. The first demonstration of lipids as second messengers was indicated by different works, which independently elucidated the process through which PI(4,5)P2 is cleaved by phospholipases C to diacyglycerol (DAG) and inositol 3-phosphate (IP3). These, in turn, contribute to protein kinases C (PKC) activation and calcium (Ca2+) release from the endoplasmic reticulum [1][2][3][4][5][7]. Subsequently, further studies have led to the discovery of many other PI-related pathways, including those involving several forms of phosphotransferases like PIKinases and PIPKinases [8] or phosphatases like phosphatase and tension homologue deleted on chromosome 10 (PTEN) and SH2-domain containing inositol phosphatase 2 (SHIP2) [9][10].
1.2. Nuclear Lipid Signalling: Focus on Nuclear Phosphoinositides
PI signaling was first described at the plasma membrane level, which involves PI anchored to cell membranes through DAG molecules [1][2][3][4][5]. Interestingly, it soon became clear that PIs and PI-related enzymes could be present in different cellular compartments, including cytoplasmic organellar membranes and nuclei. Nuclear PI fraction was expected to exist due to the presence of the nuclear membrane, a bilayer formed by lipids and proteins connected to the endoplasmic reticulum (ER) [11][12]. However, different reports have indicated that nuclei almost completely depleted of nuclear envelope are still characterized by the presence of many PIs and PI-related proteins like phospholipases C, phosphatidylinositol phosphate kinases (PIPKs) and diacylglycerol kinases (DGKs) [13][14][15][16][17][18][19]. Strikingly, under different stimuli, these enzymes are able to change the nuclear pool of PIs [14][16][20][21]. This evidence unequivocally demonstrated the existence of PI signaling completely localized within the nuclear compartment of cells.
Seven different phosphoderivatives of PI are known to exist and these are modulated by an intricate network of enzymes whose related pathways often intertwine. Despite the rarity of these second messengers when compared with other lipids like phosphatidylcholine or phosphatidylserine (which represent around the 12–20% of the cellular lipid pool) [1][2][3][4][5], they have been described as modulating many cellular functions acting in different cellular compartments.
2. Phosphatidylinositol 5 Phosphate: A Rare But Essential Lipid
2.1. PI5P Discovery
Out of the different phosphoinositides, PI5P represents only 0.5% of the PI pool present in the cells. However, its function as a second messenger has been widely investigated. Its levels strongly fluctuate due to external stimuli such as TCR activation, insulin treatment, oxidative stress, and pathogen cellular invasion [22]. PI5P was the last PI to be discovered. In the late 1980s, experiments regarding the purification of proteins involved in PI signaling indicated the existence of two related subfamilies of phosphatidylinositol phosphate kinases (PIPK) named type I and type II PIP5K. They were thought to both be able to phosphorylate PI4P, leading to PI(4,5P)2 production. However, in 1997, Rameh and colleagues, studying the substrate specificity of these two PIPK classes, found that type II enzymes were able to specifically phosphorylate another lipid substrate to lead to PI(4,5)P2, which turned out to be PI5P. As synthetic purified lipid substrates were not available at the time, the issue with PI5P detection was due to the contamination of PI4P bovine brain preparations used for experiments involving PI5P. This delayed the detection of the real function of type II PIPK, which from that point was renamed PI5P4K/PIP4K [23] (see later).
2.2. Changes in PI5P Levels Regulate Many Cellular Functions
As already stated, the pool of cellular PI5P can be regulated by many external factors and stimuli. For instance, upon TCR stimulation, quick (two minute) and transient accumulation of PI5P in Uh78 cells occurs. PI5P is in turn bound by DOK proteins, leading to IL-2 promoter activity in T cells [24]. In addition, induction of platelet aggregation by thrombin treatment has been partially connected to a three-fold increase in PI5P levels in cells [25]. Other reports have described insulin treatment as able to increase levels of PI5P in 3T3-L1 adipocytes, CHO cells stably expressing insulin receptors, and skeletal muscle cells [26]. Interestingly, insulin-dependent PI5P accumulation has been connected with GLUT4 internalization-enhancing glucose uptake from the extracellular environment [27]. On the other hand, increased levels of PI5P obtained by insulin or infection by Shigella flexeneri PI(4,5)P2 4-phosphatase IpgD lead to actin remodeling and endosome formation through TIAM1 [28]. S. flexeneri infection-related changes of PI5P have also been proposed to internalize and degrade cell surface levels of ICAM-1, inhibiting neutrophils recruitment [29]. Recently, other pathogen signatures like lipopolysaccharides (LPS) and viral dsRNA have been found to positively affect PI5P amounts in host cells, which have been described as being involved in toll-like receptor-related pathways [30]. All these reports showed that fluctuations of PI5P in cells can be linked to different external stimuli and signaling pathways.
2.3. A role for PI5P in nuclear outcomes
The role of PI5P as a second messenger has been widely investigated. In particular, many processes regulated by changes in PI5P levels have been described in relation to the nuclei. This began with evidence collected during the study of the cell cycle of murine erythroleukemia (MEL) cells, which showed a strong increase of the nuclear PI5P pool during G1/S transition [14][31]. This led to the first ideas about possible roles of this phosphoinositide in the regulation of nuclear processes [32]. In fact, throughout the years, it has turned out that PI5P is involved in many nuclear outputs such as chromatin remodeling, gene expression, or responses to stressors like UV irradiation or genotoxic factors. For example, Gozani et al. described a role for nuclear PI5P in the regulation of ING2 mediated p53 acetylation and apoptosis during stress response [33][34]. This was possible through direct interaction between ING2 and PI5P, and controlled by the activity of two different classes of enzymes named PI5P4K/PIP4K and Type I PI(4,5)P2 4-phosphatases (see later) [35][36]. Another report, indicated PI5P as able to impact on oxidative stress cellular response and reactive oxygen species (ROS) production through Pin1 [37]. Nuclear PIs are often involved in chromatin remodeling and gene expression. In particular, PI5P has been reported to negatively control Arabidopsis Thritorax (ATX) which encodes plant methylatransferase proteins, able to methylate lysine residues on the tail of H3 histones [38][39]. On the other hand, the Ubiquitin-like, containing PHD and RING Finger domains 1 (Uhrf1), an E3 ubiquitin ligase, which plays a pivotal role in DNA methyltransferase 1 (DNMT1) DNA docking, has been shown to interact with PI5P. This interaction modulates the DNA accessibility of DNMT1 and, in turn, DNA methylation [40]. Recently, we demonstrated that silencing PIP4K2B and PIP4K2C in human T regulatory cells (Treg) impact on Uhrf1 levels and Treg function and proliferation [41]. Finally, nuclear PI5P has been connected to the regulation of myogenic differentiation affecting the DNA binding to specific DNA regions of the TATA-Box Binding Protein Associated Factor 3 (TAF3), and impacting on the expression of several genes involved in myogenesis [42].
3. Enzymes Involved in the Turnover of PI5P
Although PI5P levels can change upon different stimuli, how this occurs is not always understood. Several pathways underlying PI5P synthesis have been described, including direct processes through phosphorylation of PI on position 5, or indirect-like de-phosphorylation of PI(3,5)P2 (Figure 1). Moreover, the balance between PI(4,5)P2 and PI5P mediated by PIP4K has also been found to be important for cellular control of the PI5P pool.
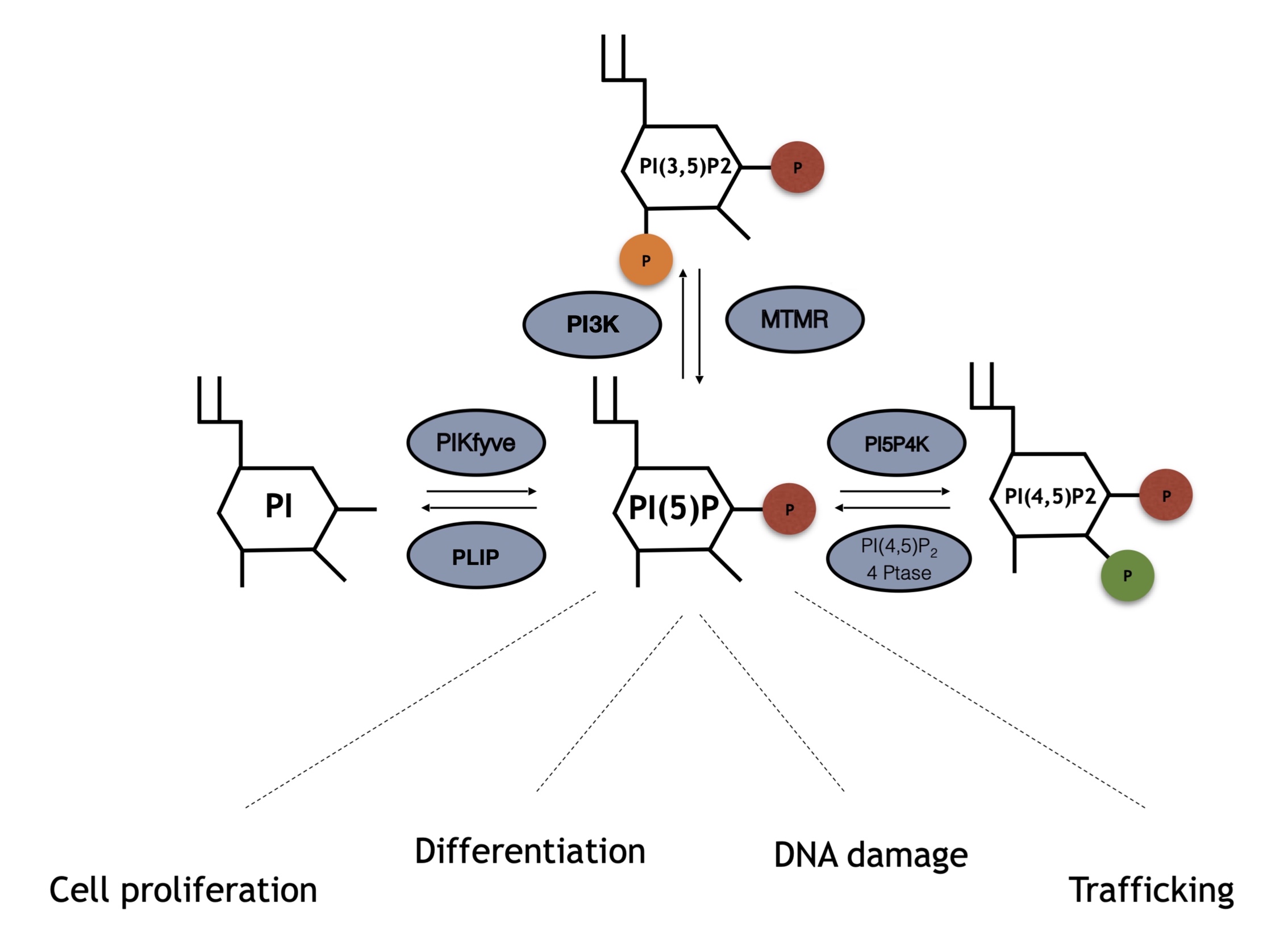
Figure 1. Enzymes involved in phosphatidylinositol 5 phosphate (PI5P) turnover and array of kinases and phosphatases involved in PI5P turnover. PI5P can be directly synthesized by PIKFyve phosphotransferases through direct phosphorylation of phosphatidylinositol (PI) on position 5 of the inositol ring. Moreover, MTMR phosphatases can remove a phosphate group on position 3 from PI(3,5)P2, leading to increased amount of PI5P levels. Finally, PI5P4K/PIP4Ks directly phosphorylate PI5P on position 4 leading to PI(4,5)P2 synthesis, an event that can be counterbalanced by type I/II 4-phosphatases which remove the phosphate group on position 4.
3.1. PIKfyve/Phosphatidylinositol-3-Phosphate 5-Kinase
PIKfyve, also known as phosphatidylinositol-3-phosphate 5-kinase type III or PIPKIII, is an established evolutionarily conserved PIK present in animals, plants and fungi. It possesses a FYVE zinc finger domain, named after the proteins in which it was first identified: Fab1p (the yeast orthologue of PIKfyve), YOTB, Vac1 (vesicle transport protein), and EEA1 (Early Endosome Antigen 1) [43][44][45][46]. This domain has a small cysteine-rich Zn2+ binding domain, characterized by a basic motif in the first β-strand (R/K) (R/K) HHCR which primarily allows phosphatidylinositol 3 phosphate (PI3P) binding. PIKfyve is a large protein involved in endosome processing, HIV and Salmonella replication, and type 2 diabetes, while mutations in its coding gene are connected to corneal fleck dystrophy (CFD) [46][47]. Interestingly, together with its capacity to phosphorylate PIs, it possesses protein kinase activity towards non lipid substrates [48]. Nevertheless, in vitro and in vivo evidence has described PIKfyve as able to bind and phosphorylate the lipids PI3P and PI, leading to the synthesis of PI(3,5)P2 and PI5P respectively [43][44][45][46] (Figure 1). Overexpression or silencing/inhibition of PIKfyve leads to changes in the levels of these two PIs. Most PI5P production in cells is thought to be due to PIKfyve activity via both direct and indirect pathways. Indeed, as stated, PIKfyve is able to directly phosphorylate PI rings in position 5, leading to synthesis of PI5P (the direct pathway) [43][44][45][46][49]. Another proposed model of PI5P synthesis is related to dephosphorylation of PIKfyve-derived PI(3,5)P2 by 3-phosphatases named myotubularins (the indirect pathway, see next) [50][51].
3.2. MTM-MTMR/Myotubularins
Myotubularin 3-phosphatases represent a family of proteins conserved in eukaryotes and composed of 15 members named MTM1 and MTMR1–14 [52][53][54][55]. These enzymes share a structural motif which is represented by a PH-GRAM (pleckstrin homology-glucosyltransferases, rab-like GTPase activators, and myotubularin), catalytic protein tyrosine phosphatases (PTP) domains, and a coiled-coil motif. Some of the isoforms also contain FYVE-, PH-, and PDZ-binding sites [52][53][54][55]. The active site of the protein is represented by a Cys-X5-Arg motif, which allows hydrolyzation of phosphodiester bonds on a cysteine nucleophile and an arginine residue, binding an oxygen atom onto the phosphate groups [52][53][54][55]. These catalytic residues can be altered by missense substitutions in several isoforms. This divides myotubularins into active (MTM1, MTMR1–4, MTMR6, MTMR7–8, and MTMR14) and inactive (MTMR5 and MTMR9–13) phosphatases [56][57]. Throughout the years, MTMs have been described as being able to bind and dephosphorylate PI3P and PI(3,5)P2 to PI and PI5P, respectively. Reports on the crystal structure of MTMR2 have unraveled the molecular basis of PI3P and PI(3,5)P2 binding through its PH-GRAM domain [57][58][59] (Figure 1). Finally, this class of PI phosphatases is known to play a role in endocytosis and membrane trafficking, cell proliferation, differentiation, and cell junction dynamics. Mutations on the genes encoding myotubularin enzymes have been found in neuromuscular diseases or have been associated with metabolic syndromes, obesity, and cancer.
3.3. Phosphatidylinositol 5 Phosphate 4 Kinases (PI5P4K/PIP4K)
Phosphatidylinositol 5 phosphate 4 kinases, or type II PIPKs, represent a family of enzymes able to phosphorylate PI5P in order to produce PI(4,5)P2 [23]. PIP4Ks are conserved in different species spanning from flies and worms to mice and humans. Mammalians are characterized by the presence of three different isoforms, namely, PIP4K2A, PIP4K2B, and PIP4K2C [4][8][60][61][62]. They share dimerization and lipid kinase domains and carry main differences in amino acid sequences found at -N and -C termini, which confer each isoform specific characteristics [4][8][61][62][63]. PIP4K2A is considered the most active isozyme if compared to PIP4K2B, while PIP4K2C possesses a limited capacity to phosphorylate PI5P [4][8][60][61][62]. Interestingly, PIP4K2B preferentially uses GTP instead of ATP for its kinase function [64][65]. PIP4K isozymes also differ from each other for their intracellular localization: 2A is mostly located in the cytoplasm/membrane, 2B can also be found in the nucleus, and 2C is found in not well defined membraneless compartments. As already indicated, this class of enzymes was first discovered in 1997 by members of Cantley’s lab, who were able to overcome an issue related to the mix between PI4P and PI5P in bovine brain preparations [23]. This finding rendered the study of PIP4K substrate specificity possible and described those proteins as different from their related family of PIP5K [66]. In any case, the capacity of PIP4K to produce PI(4,5)P2 is considered minor with respect to PIP5K, so they are suggested as being involved in the regulation of PI5P levels in cells [4][8][61][62] (Figure 1). Indeed, knockdown/inhibition of PIP4Ks in mammalian cells or knockout in drosophila leads to increased levels of PI5P, with almost no effects on the pool of PI(4,5)P2 [61][67][68]. These proteins have been recently connected with many cellular functions, including DNA damage, cell proliferation, and chromatin remodeling, and have been proposed as possible targets for treatment of cancer or autoimmune diseases [69][70][71][72][41][73].
3.4. Type I/II PI(4,5)P2 4-Phosphatases
Type I/II PI(4,5)P2 4-phosphatases are human phosphatases able to specifically target PI(4,5)P2 and hydrolyze the phosphodiester bond of D4 phosphate on the inositol ring, leading to synthesis of PI5P in human cells. Type I/II 4-phosphatases share a Cys-X5-Arg motif with the Shigella flexenery PI(4,5)P2 4-phosphatase IpgD. This, once injected or expressed into host cells, leads to a specific and strong increase in PI5P levels through PI(4,5)P2 hydrolysis [74][75][76] (Figure 1). Mammalian type I/II PI(4,5)P2 4-phosphatases were described for the first time by Ungewickell et al. and were reported as being located in endosomal/lysosomal membranes in epithelial cells. In addition, upon cellular stress, type I 4-phosphatase can localize in the nucleus, where it regulates p53 dependent apoptosis [35].
4. Conclusions
Despite the low abundance in the cells, PI5P is a lipid second messenger involved in the regulation of several cellular functions, spanning from cytoskeletal organisation to nuclear dynamics. Specifically, PI5P production and turnover is due to the activity of different enzymes comprising myotubularins, PIKFyve, PIP4Ks, and type I/II 4-phosphatases. However, the study of PI5P as a second messenger is still quite challenging. This is due to a lack of specific antibodies or ways to detect it, i.e., more sensitive cellular probes, as well as methods to specifically and singularly alter its levels. For all these reasons, many new chapters in the story of this rare and little understood phosphoinositide will be written in future.
References
- Berridge, M.J.; Irvine, R.F. Inositol phosphates and cell signalling. Nature 1989, 341, 197–205.
- Di Paolo, G.; De Camilli, P. Phosphoinositides in cell regulation and membrane dynamics. Nature 2006, 443, 651–657.
- Wymann, M.P.; Schneiter, R. Lipid signalling in disease. Nat. Rev. Mol. Cell. Biol. 2008, 9, 162–176.
- Poli, A.; Billi, A.M.; Mongiorgi, S.; Ratti, S.; McCubrey, J.A.; Suh, P.G.; Cocco, L.; Ramazzotti, G. Nuclear Phosphatidylinositol Signaling: Focus on Phosphatidylinositol Phosphate Kinases and Phospholipases, C. J. Cell. Physiol. 2016, 231, 1645–1655.
- Dickson, E.J.; Hille, B. Understanding phosphoinositides: Rare, dynamic, and essential membrane phospholipids. Biochem. J. 2019, 476, 1–23.
- Epand, R.M. Features of the Phosphatidylinositol Cycle and its Role in Signal Transduction. J. Membr. Biol. 2017, 250, 353–366.
- Newton, A.C. Protein kinase C: Structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem. Rev. 2001, 101, 2353–2364.
- Burke, J.E. Structural Basis for Regulation of Phosphoinositide Kinases and Their Involvement in Human Disease. Mol. Cell 2018, 71, 653–673.
- Simpson, L.; Parsons, R. PTEN: Life as a tumor suppressor. Exp. Cell Res. 2001, 264, 29–41.
- Erneux, C.; Edimo, W.E.; Deneubourg, L.; Pirson, I. SHIP2 multiple functions: A balance between a negative control of PtdIns(3,4,5)P(3) level, a positive control of PtdIns(3,4)P(2) production, and intrinsic docking properties. J. Cell. Biochem. 2011, 112, 2203–2209.
- Smith, C.D.; Wells, W.W. Phosphorylation of rat liver nuclear envelopes. II. Characterization of in vitro lipid phosphorylation. J. Biol. Chem. 1983, 258, 9368–9373.
- van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell. Biol. 2008, 9, 112–124.
- Cocco, L.; Gilmour, R.S.; Ognibene, A.; Letcher, A.J.; Manzoli, F.A.; Irvine, R.F. Synthesis of polyphosphoinositides in nuclei of Friend cells. Evidence for polyphosphoinositide metabolism inside the nucleus which changes with cell differentiation. Biochem. J. 1987, 248, 765–770.
- Divecha, N.; Banfic, H.; Irvine, R.F. The polyphosphoinositide cycle exists in the nuclei of Swiss 3T3 cells under the control of a receptor (for IGF-I) in the plasma membrane, and stimulation of the cycle increases nuclear diacylglycerol and apparently induces translocation of protein kinase C to the nucleus. EMBO J. 1991, 10, 3207–3214.
- Payrastre, B.; Nievers, M.; Boonstra, J.; Breton, M.; Verkleij, A.J.; Van Bergen en Henegouwen, P.M. A differential location of phosphoinositide kinases, diacylglycerol kinase, and phospholipase C in the nuclear matrix. J. Biol. Chem. 1992, 267, 5078–5084.
- Vann, L.R.; Wooding, F.B.; Irvine, R.F.; Divecha, N. Metabolism and possible compartmentalization of inositol lipids in isolated rat-liver nuclei. Biochem. J. 1997, 327, 569–576.
- Irvine, R.F. Nuclear lipid signalling. Nat. Rev. Mol. Cell. Biol. 2003, 4, 349–360.
- Merida, I.; Avila-Flores, A.; Merino, E. Diacylglycerol kinases: At the hub of cell signalling. Biochem. J. 2008, 409, 1–18.
- Suh, P.G.; Park, J.I.; Manzoli, L.; Cocco, L.; Peak, J.C.; Katan, M.; Fukami, K.; Kataoka, T.; Yun, S.; Ryu, S.H. Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Rep. 2008, 41, 415–434.
- Capitani, S.; Cocco, L.; Maraldi, N.M.; Mazzotti, G.; Barnabei, O.; Manzoli, F.A. Inositol lipid phosphorylation in the cell nucleus. Adv. Enzym. Regul. 1991, 31, 399–416.
- Divecha, N.; Letcher, A.J.; Banfic, H.H.; Rhee, S.G.; Irvine, R.F. Changes in the components of a nuclear inositide cycle during differentiation in murine erythroleukaemia cells. Biochem. J. 1995, 312, 63–67.
- Hasegawa, J.; Strunk, B.S.; Weisman, L.S. PI5P and PI(3,5)P2: Minor, but Essential Phosphoinositides. Cell Struct. Funct. 2017, 42, 49–60.
- Rameh, L.E.; Tolias, K.F.; Duckworth, B.C.; Cantley, L.C. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature 1997, 390, 192–196.
- Nunes, J.A.; Guittard, G. An Emerging Role for PI5P in T Cell Biology. Front. Immunol. 2013, 4, 80.
- Morris, J.B.; Hinchliffe, K.A.; Ciruela, A.; Letcher, A.J.; Irvine, R.F. Thrombin stimulation of platelets causes an increase in phosphatidylinositol 5-phosphate revealed by mass assay. FEBS Lett. 2000, 475, 57–60.
- Coronas, S.; Ramel, D.; Pendaries, C.; Gaits-Iacovoni, F.; Tronchere, H.; Payrastre, B. PtdIns5P: A little phosphoinositide with big functions? Biochem. Soc. Symp. 2007, 74, 117–128.
- Grainger, D.L.; Tavelis, C.; Ryan, A.J.; Hinchliffe, K.A. Involvement of phosphatidylinositol 5-phosphate in insulin-stimulated glucose uptake in the L6 myotube model of skeletal muscle. Pflug. Arch. Eur. J. Physiol. 2011, 462, 723–732.
- Viaud, J.; Lagarrigue, F.; Ramel, D.; Allart, S.; Chicanne, G.; Ceccato, L.; Courilleau, D.; Xuereb, J.M.; Pertz, O.; Payrastre, B.; et al. Phosphatidylinositol 5-phosphate regulates invasion through binding and activation of Tiam1. Nat. Commun. 2014, 5, 4080.
- Boal, F.; Puhar, A.; Xuereb, J.M.; Kunduzova, O.; Sansonetti, P.J.; Payrastre, B.; Tronchere, H. PI5P Triggers ICAM-1 Degradation in Shigella Infected Cells, Thus Dampening Immune Cell Recruitment. Cell Rep. 2016, 14, 750–759.
- Kawasaki, T.; Takemura, N.; Standley, D.M.; Akira, S.; Kawai, T. The second messenger phosphatidylinositol-5-phosphate facilitates antiviral innate immune signaling. Cell Host Microbe 2013, 14, 148–158.
- Divecha, N.; Banfic, H.; Irvine, R.F. Inositides and the nucleus and inositides in the nucleus. Cell 1993, 74, 405–407.
- Gong, W.; Suzuki, K.; Russell, M.; Riabowol, K. Function of the ING family of PHD proteins in cancer. Int. J. Biochem. Cell Biol. 2005, 37, 1054–1065.
- Gozani, O.; Karuman, P.; Jones, D.R.; Ivanov, D.; Cha, J.; Lugovskoy, A.A.; Baird, C.L.; Zhu, H.; Field, S.J.; Lessnick, S.L.; et al. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 2003, 114, 99–111.
- Li, X.; Kikuchi, K.; Takano, Y. ING Genes Work as Tumor Suppressor Genes in the Carcinogenesis of Head and Neck Squamous Cell Carcinoma. J. Oncol. 2011, 2011, 963614.
- Zou, J.; Marjanovic, J.; Kisseleva, M.V.; Wilson, M.; Majerus, P.W. Type I phosphatidylinositol-4,5-bisphosphate 4-phosphatase regulates stress-induced apoptosis. Proc. Natl. Acad. Sci. USA 2007, 104, 16834–16839.
- Jones, D.R.; Bultsma, Y.; Keune, W.J.; Halstead, J.R.; Elouarrat, D.; Mohammed, S.; Heck, A.J.; D’Santos, C.S.; Divecha, N. Nuclear PtdIns5P as a transducer of stress signaling: An in vivo role for PIP4Kbeta. Mol. Cell 2006, 23, 685–695.
- Bua, D.J.; Martin, G.M.; Binda, O.; Gozani, O. Nuclear phosphatidylinositol-5-phosphate regulates ING2 stability at discrete chromatin targets in response to DNA damage. Sci. Rep. 2013, 3, 2137.
- Alvarez-Venegas, R.; Sadder, M.; Hlavacka, A.; Baluska, F.; Xia, Y.; Lu, G.; Firsov, A.; Sarath, G.; Moriyama, H.; Dubrovsky, J.G.; et al. The Arabidopsis homolog of trithorax, ATX1, binds phosphatidylinositol 5-phosphate, and the two regulate a common set of target genes. Proc. Natl. Acad. Sci. USA 2006, 103, 6049–6054.
- Alvarez-Venegas, R.; Pien, S.; Sadder, M.; Witmer, X.; Grossniklaus, U.; Avramova, Z. ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr. Biol. 2003, 13, 627–637.
- Gelato, K.A.; Tauber, M.; Ong, M.S.; Winter, S.; Hiragami-Hamada, K.; Sindlinger, J.; Lemak, A.; Bultsma, Y.; Houliston, S.; Schwarzer, D.; et al. Accessibility of different histone H3-binding domains of UHRF1 is allosterically regulated by phosphatidylinositol 5-phosphate. Mol. Cell 2014, 54, 905–919.
- Poli A., Abdul-Hamidc S., Zaurito A., Campagnoli F., Bevilacqua A., Sheth B., Fiume R., Pagani M., Abrignani S., and Divecha N.; PIP4Ks impact on PI3K, FOXP3, and UHRF1 signaling and modulate human regulatory T cell proliferation and immunosuppressive activity. Proceedings of the National Academy of Sciences of the United States of America (PNAS) 2021, Vol. 118 No. 31 , 0, 10.1073/pnas.2010053118.
- Stijf-Bultsma, Y.; Sommer, L.; Tauber, M.; Baalbaki, M.; Giardoglou, P.; Jones, D.R.; Gelato, K.A.; van Pelt, J.; Shah, Z.; Rahnamoun, H.; et al. The basal transcription complex component TAF3 transduces changes in nuclear phosphoinositides into transcriptional output. Mol. Cell 2015, 58, 453–467.
- Sbrissa, D.; Ikonomov, O.C.; Shisheva, A. PIKfyve, a mammalian ortholog of yeast Fab1p lipid kinase, synthesizes 5-phosphoinositides. Effect of insulin. J. Biol. Chem. 1999, 274, 21589–21597.
- Shisheva, A.; Sbrissa, D.; Ikonomov, O. Cloning, characterization, and expression of a novel Zn2+-binding FYVE finger-containing phosphoinositide kinase in insulin-sensitive cells. Mol. Cell. Biol. 1999, 19, 623–634.
- Sbrissa, D.; Ikonomov, O.C.; Deeb, R.; Shisheva, A. Phosphatidylinositol 5-phosphate biosynthesis is linked to PIKfyve and is involved in osmotic response pathway in mammalian cells. J. Biol. Chem. 2002, 277, 47276–47284.
- Shisheva, A. PIKfyve and its Lipid products in health and in sickness. Curr. Top. Microbiol. Immunol. 2012, 362, 127–162.
- Sbrissa, D.; Ikonomov, O.C.; Filios, C.; Delvecchio, K.; Shisheva, A. Functional dissociation between PIKfyve-synthesized PtdIns5P and PtdIns(3,5)P2 by means of the PIKfyve inhibitor YM201636. Am. J. Physiol. Cell Physiol. 2012, 303, C436–C446.
- Sbrissa, D.; Ikonomov, O.C.; Shisheva, A. PIKfyve lipid kinase is a protein kinase: Downregulation of 5’-phosphoinositide product formation by autophosphorylation. Biochemistry 2000, 39, 15980–15989.
- Shisheva, A.; Sbrissa, D.; Ikonomov, O. Plentiful PtdIns5P from scanty PtdIns(3,5)P2 or from ample PtdIns? PIKfyve-dependent models: Evidence and speculation (response to: DOI 10.1002/bies.201300012). Bioessays 2015, 37, 267–277.
- Vaccari, I.; Dina, G.; Tronchere, H.; Kaufman, E.; Chicanne, G.; Cerri, F.; Wrabetz, L.; Payrastre, B.; Quattrini, A.; Weisman, L.S.; et al. Genetic interaction between MTMR2 and FIG4 phospholipid phosphatases involved in Charcot-Marie-Tooth neuropathies. PLoS Genet. 2011, 7, e1002319.
- Oppelt, A.; Lobert, V.H.; Haglund, K.; Mackey, A.M.; Rameh, L.E.; Liestol, K.; Schink, K.O.; Pedersen, N.M.; Wenzel, E.M.; Haugsten, E.M.; et al. Production of phosphatidylinositol 5-phosphate via PIKfyve and MTMR3 regulates cell migration. EMBO Rep. 2013, 14, 57–64.
- Blondeau, F.; Laporte, J.; Bodin, S.; Superti-Furga, G.; Payrastre, B.; Mandel, J.L. Myotubularin, a phosphatase deficient in myotubular myopathy, acts on phosphatidylinositol 3-kinase and phosphatidylinositol 3-phosphate pathway. Hum. Mol. Genet. 2000, 9, 2223–2229.
- Taylor, G.S.; Maehama, T.; Dixon, J.E. Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc. Natl. Acad. Sci. USA 2000, 97, 8910–8915.
- Kim, S.A.; Taylor, G.S.; Torgersen, K.M.; Dixon, J.E. Myotubularin and MTMR2, phosphatidylinositol 3-phosphatases mutated in myotubular myopathy and type 4B Charcot-Marie-Tooth disease. J. Biol. Chem. 2002, 277, 4526–4531.
- Schaletzky, J.; Dove, S.K.; Short, B.; Lorenzo, O.; Clague, M.J.; Barr, F.A. Phosphatidylinositol-5-phosphate activation and conserved substrate specificity of the myotubularin phosphatidylinositol 3-phosphatases. Curr. Biol. 2003, 13, 504–509.
- Doerks, T.; Strauss, M.; Brendel, M.; Bork, P. GRAM, a novel domain in glucosyltransferases, myotubularins and other putative membrane-associated proteins. Trends. Biochem. Sci. 2000, 25, 483–485.
- Begley, M.J.; Dixon, J.E. The structure and regulation of myotubularin phosphatases. Curr. Opin. Struct. Biol. 2005, 15, 614–620.
- Begley, M.J.; Taylor, G.S.; Kim, S.A.; Veine, D.M.; Dixon, J.E.; Stuckey, J.A. Crystal structure of a phosphoinositide phosphatase, MTMR2: Insights into myotubular myopathy and Charcot-Marie-Tooth syndrome. Mol. Cell 2003, 12, 1391–1402.
- Begley, M.J.; Taylor, G.S.; Brock, M.A.; Ghosh, P.; Woods, V.L.; Dixon, J.E. Molecular basis for substrate recognition by MTMR2, a myotubularin family phosphoinositide phosphatase. Proc. Natl. Acad. Sci. USA 2006, 103, 927–932.
- Clarke, J.H.; Irvine, R.F. Evolutionarily conserved structural changes in phosphatidylinositol 5-phosphate 4-kinase (PI5P4K) isoforms are responsible for differences in enzyme activity and localization. Biochem. J. 2013, 454, 49–57.
- Bulley, S.J.; Clarke, J.H.; Droubi, A.; Giudici, M.L.; Irvine, R.F. Exploring phosphatidylinositol 5-phosphate 4-kinase function. Adv. Biol. Regul. 2015, 57, 193–202.
- Fiume, R.; Stijf-Bultsma, Y.; Shah, Z.H.; Keune, W.J.; Jones, D.R.; Jude, J.G.; Divecha, N. PIP4K and the role of nuclear phosphoinositides in tumour suppression. Biochim. Biophys. Acta 2015, 1851, 898–910.
- Rao, V.D.; Misra, S.; Boronenkov, I.V.; Anderson, R.A.; Hurley, J.H. Structure of type IIbeta phosphatidylinositol phosphate kinase: A protein kinase fold flattened for interfacial phosphorylation. Cell 1998, 94, 829–839.
- Fiume, R.; Jones, D.R.; Divecha, N. PIP4K2B: Coupling GTP Sensing to PtdIns5P Levels to Regulate Tumorigenesis. Trends Biochem. Sci. 2016, 41, 473–475.
- Sumita, K.; Lo, Y.H.; Takeuchi, K.; Senda, M.; Kofuji, S.; Ikeda, Y.; Terakawa, J.; Sasaki, M.; Yoshino, H.; Majd, N.; et al. The Lipid Kinase PI5P4Kbeta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis. Mol. Cell 2016, 61, 187–198.
- van den Bout, I.; Divecha, N. PIP5K-driven PtdIns(4,5)P2 synthesis: Regulation and cellular functions. J. Cell Sci. 2009, 122, 3837–3850.
- Al-Ramahi, I.; Giridharan, S.S.P.; Chen, Y.C.; Patnaik, S.; Safren, N.; Hasegawa, J.; de Haro, M.; Wagner Gee, A.K.; Titus, S.A.; Jeong, H.; et al. Inhibition of PIP4Kgamma ameliorates the pathological effects of mutant huntingtin protein. Elife 2017, 6, e29123.
- Lundquist, M.R.; Goncalves, M.D.; Loughran, R.M.; Possik, E.; Vijayaraghavan, T.; Yang, A.; Pauli, C.; Ravi, A.; Verma, A.; Yang, Z.; et al. Phosphatidylinositol-5-Phosphate 4-Kinases Regulate Cellular Lipid Metabolism By Facilitating Autophagy. Mol. Cell 2018, 70, 531–544.
- Emerling, B.M.; Hurov, J.B.; Poulogiannis, G.; Tsukazawa, K.S.; Choo-Wing, R.; Wulf, G.M.; Bell, E.L.; Shim, H.S.; Lamia, K.A.; Rameh, L.E.; et al. Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell 2013, 155, 844–857.
- Keune, W.J.; Sims, A.H.; Jones, D.R.; Bultsma, Y.; Lynch, J.T.; Jirstrom, K.; Landberg, G.; Divecha, N. Low PIP4K2B expression in human breast tumors correlates with reduced patient survival: A role for PIP4K2B in the regulation of E-cadherin expression. Cancer Res. 2013, 73, 6913–6925.
- Jude, J.G.; Spencer, G.J.; Huang, X.; Somerville, T.D.D.; Jones, D.R.; Divecha, N.; Somervaille, T.C.P. A targeted knockdown screen of genes coding for phosphoinositide modulators identifies PIP4K2A as required for acute myeloid leukemia cell proliferation and survival. Oncogene 2015, 34, 1253–1262.
- Shim, H.; Wu, C.; Ramsamooj, S.; Bosch, K.N.; Chen, Z.; Emerling, B.M.; Yun, J.; Liu, H.; Choo-Wing, R.; Yang, Z.; et al. Deletion of the gene Pip4k2c, a novel phosphatidylinositol kinase, results in hyperactivation of the immune system. Proc. Natl. Acad. Sci. USA 2016, 113, 7596–7601.
- Chen S., Tjin C. C., Gao X., Xue Y., Jiao H., Zhang R., Wu M., He Z., Ellman J., and Ha Y.; Pharmacological inhibition of PI5P4Kα/β disrupts cell energy metabolism and selectively kills p53-null tumor cells. Proceedings of the National Academy of Sciences of the United States of America 2021, Vol. 118 (21) , 0, 10.1073/pnas.2002486118.
- Niebuhr, K.; Giuriato, S.; Pedron, T.; Philpott, D.J.; Gaits, F.; Sable, J.; Sheetz, M.P.; Parsot, C.; Sansonetti, P.J.; Payrastre, B. Conversion of PtdIns(4,5)P(2) into PtdIns(5)P by the S.flexneri effector IpgD reorganizes host cell morphology. EMBO J. 2002, 21, 5069–5078.
- Ungewickell, A.; Hugge, C.; Kisseleva, M.; Chang, S.C.; Zou, J.; Feng, Y.; Galyov, E.E.; Wilson, M.; Majerus, P.W. The identification and characterization of two phosphatidylinositol-4,5-bisphosphate 4-phosphatases. Proc. Natl. Acad. Sci. USA 2005, 102, 18854–18859.
- Mason, D.; Mallo, G.V.; Terebiznik, M.R.; Payrastre, B.; Finlay, B.B.; Brumell, J.H.; Rameh, L.; Grinstein, S. Alteration of epithelial structure and function associated with PtdIns(4,5)P2 degradation by a bacterial phosphatase. J. Gen. Physiol. 2007, 129, 267–283.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Revisions:
2 times
(View History)
Update Date:
11 Aug 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




