
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Paula Nunes-Hasler | + 3301 word(s) | 3301 | 2021-06-10 06:22:46 | | | |
| 2 | Vivi Li | Meta information modification | 3301 | 2021-07-15 03:49:13 | | |
Video Upload Options
Oxysterol binding related proteins 5 and 8 (ORP5 and ORP8) are two close homologs of the larger oxysterol binding protein (OSBP) family of sterol sensors and lipid transfer proteins (LTP). Early studies indicated these transmembrane proteins, anchored to the endoplasmic reticulum (ER), bound and sensed cholesterol and oxysterols. They were identified as important for diverse cellular functions including sterol homeostasis, vesicular trafficking, proliferation and migration. In addition, they were implicated in lipid-related diseases such as atherosclerosis and diabetes, but also cancer, although their mechanisms of action remained poorly understood. Then, alongside the increasing recognition that membrane contact sites (MCS) serve as hubs for non-vesicular lipid transfer, added to their structural similarity to other LTPs, came discoveries showing that ORP5 and 8 were in fact phospholipid transfer proteins that rather sense and exchange phosphatidylserine (PS) for phosphoinositides, including phosphatidylinositol-4-phosphate (PI(4)P) and potentially phosphatidylinositol-(4,5)-bisphosphate (PI(4,5)P2). Evidence now points to their action at MCS between the ER and various organelles including the plasma membrane, lysosomes, mitochondria, and lipid droplets. Dissecting exactly how this unexpected phospholipid transfer function connects with sterol regulation in health or disease remains a challenge for future studies.
1. Introduction
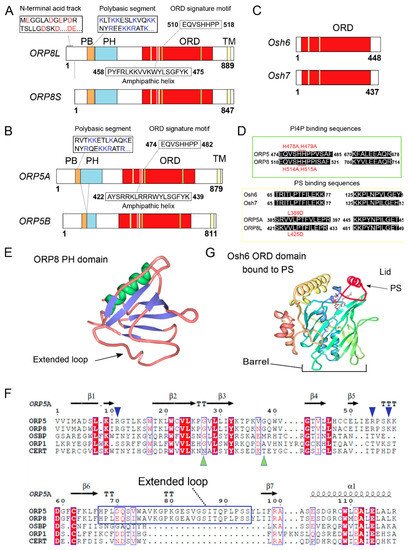
2. ORP5/8 as Oxysterol Sensors
3. ORP5/8 in the Control of Vesicular Trafficking
References
- Munro, S. Organelle identity and the organization of membrane traffic. Nature 2004, 6, 469–472.
- Holthuis, J.C.; Menon, A.K. Lipid landscapes and pipelines in membrane homeostasis. Nature 2014, 510, 48–57.
- Lorizate, M.; Kräusslich, H.-G. Role of Lipids in Virus Replication. Cold Spring Harb. Perspect. Boil. 2011, 3, a004820.
- A Kandutsch, A.; Shown, E.P. Assay of oxysterol-binding protein in a mouse fibroblast, cell-free system. Dissociation constant and other properties of the system. J. Boil. Chem. 1981, 256, 13068–13073.
- Taylor, F.R.; Kandutsch, A.A. Oxysterol binding protein. Chem. Phys. Lipids 1985, 38, 187–194.
- Pietrangelo, A.; Ridgway, N.D. Bridging the molecular and biological functions of the oxysterol-binding protein family. Cell. Mol. Life Sci. 2018, 75, 3079–3098.
- Jaworski, C.; Moreira, E.; Li, A.; Lee, R.; Rodriguez, I.R. A Family of 12 Human Genes Containing Oxysterol-Binding Domains. Genomics 2001, 78, 185–196.
- Laitinen, S.; Olkkonen, V.M.; Ehnholm, C.; Ikonen, E. Family of human oxysterol binding protein (OSBP) homologues. A novel member implicated in brain sterol metabolism. J. Lipid Res. 1999, 40.
- Lehto, M.; Laitinen, S.; Chinetti, G.; Johansson, M.; Ehnholm, C.; Staels, B.; Ikonen, E.; Olkkonen, V.M. The OSBP-related protein family in humans. J. Lipid Res. 2001, 42, 1203–1213.
- Lehto, M.; Olkkonen, V.M. The OSBP-related proteins: A novel protein family involved in vesicle transport, cellular lipid metabolism, and cell signalling. Biochim. Biophys. Acta (BBA) - Mol. Cell Boil. Lipids 2003, 1631, 1–11.
- Beh, C.T.; Cool, L.; Phillips, J.; Rine, J. Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics 2001, 157, 1117–1140.
- Schroepfer, G.J. Oxysterols: Modulators of cholesterol metabolism and other processes. Physiol. Rev. 2000, 80, 361–554.
- Olkkonen, V.M.; Levine, T.P. Oxysterol binding proteins: in more than one place at one time? Biochem. Cell Boil. 2004, 82, 87–98.
- Helle, S.C.; Kanfer, G.; Kolar, K.; Lang, A.; Michel, A.H.; Kornmann, B. Organization and function of membrane contact sites. Biochim. Biophys. Acta (BBA) - Bioenerg. 2013, 1833, 2526–2541.
- Jain, A.; Holthuis, J.C. Membrane contact sites, ancient and central hubs of cellular lipid logistics. Biochim. Biophys. Acta (BBA) - Bioenerg. 2017, 1864, 1450–1458.
- Ridgway, N.D.; Dawson, P.A.; Ho, Y.K.; Brown, M.S.; Goldstein, J.L. Translocation of oxysterol binding protein to Golgi apparatus triggered by ligand binding. J. Cell Boil. 1992, 116, 307–319.
- Im, Y.J.; Raychaudhuri, S.; Prinz, W.A.; Hurley, J.H. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature 2005, 437, 154–158.
- Raychaudhuri, S.; Im, Y.J.; Hurley, J.H.; Prinz, W.A. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein–related proteins and phosphoinositides. J. Cell Boil. 2006, 173, 107–119.
- De Saint-Jean, M.; Delfosse, V.; Douguet, D.; Chicanne, G.; Payrastre, B.; Bourguet, W.; Antonny, B.; Drin, G. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J. Cell Boil. 2011, 195, 965–978.
- Mesmin, B.; Bigay, J.; Von Filseck, J.M.; Lacas-Gervais, S.; Drin, G.; Antonny, B. A Four-Step Cycle Driven by PI(4)P Hydrolysis Directs Sterol/PI(4)P Exchange by the ER-Golgi Tether OSBP. Cell 2013, 155, 830–843.
- Von Filseck, J.M.; Vanni, S.; Mesmin, B.; Antonny, B.; Drin, G. A phosphatidylinositol-4-phosphate powered exchange mechanism to create a lipid gradient between membranes. Nat. Commun. 2015, 6, 6671.
- Du, X.; Kumar, J.; Ferguson, C.; Schulz, T.A.; Ong, Y.S.; Hong, W.; Prinz, W.A.; Parton, R.; Brown, A.J.; Yang, H. A role for oxysterol-binding protein–related protein 5 in endosomal cholesterol trafficking. J. Cell Boil. 2011, 192, 121–135.
- Chung, J.; Torta, F.; Masai, K.; Lucast, L.; Czapla, H.; Tanner, L.B.; Narayanaswamy, P.; Wenk, M.R.; Nakatsu, F.; De Camilli, P. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science 2015, 349, 428–432.
- Sohn, M.; Korzeniowski, M.; Zewe, J.P.; Wills, R.C.; Hammond, G.R.V.; Humpolíčková, J.; Vrzal, L.; Chalupska, D.; Veverka, V.; Fairn, G.D.; et al. PI(4,5)P2 controls plasma membrane PI4P and PS levels via ORP5/8 recruitment to ER–PM contact sites. J. Cell Boil. 2018, 217, 1797–1813.
- Lee, M.; Fairn, G.D. Both the PH domain and N-terminal region of oxysterol-binding protein related protein 8S are required for localization to PM-ER contact sites. Biochem. Biophys. Res. Commun. 2018, 496, 1088–1094.
- Ghai, R.; Du, X.; Wang, H.; Dong, J.; Ferguson, C.; Brown, A.J.; Parton, R.; Wu, J.-W.; Yang, H. ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P2) and regulate its level at the plasma membrane. Nat. Commun. 2017, 8, 757.
- Raychaudhuri, S.; Prinz, W.A. The diverse functions of oxysterol-binding proteins. Annu. Rev. Cell Dev. Boil. 2010, 26, 157–177.
- Yan, D.; Mäyränpää, M.I.; Wong, J.; Perttilä, J.; Lehto, M.; Jauhiainen, M.; Kovanen, P.T.; Ehnholm, C.; Brown, A.J.; Olkkonen, V.M. OSBP-related Protein 8 (ORP8) SuppressesABCA1Expression and Cholesterol Efflux from Macrophages. J. Boil. Chem. 2007, 283, 332–340.
- Koga, Y.; Ishikawa, S.; Nakamura, T.; Masuda, T.; Nagai, Y.; Takamori, H.; Hirota, M.; Kanemitsu, K.; Baba, Y.; Baba, H. Oxysterol binding protein-related protein-5 is related to invasion and poor prognosis in pancreatic cancer. Cancer Sci. 2008, 99, 2387–2394.
- Jordan, S.D.; Krüger, M.; Willmes, D.M.; Redemann, N.; Wunderlich, F.T.; Brönneke, H.S.; Merkwirth, C.; Kashkar, H.; Olkkonen, V.M.; Böttger, T.; et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nature 2011, 13, 434–446.
- Béaslas, O.; Metso, J.; Nissila, E.; Laurila, P.-P.; Kaiharju, E.; Batchu, K.C.; Kaipiainen, L.; Mäyränpää, M.I.; Yan, D.; Gylling, H.; et al. Osbpl8 Deficiency in Mouse Causes an Elevation of High-Density Lipoproteins and Gender-Specific Alterations of Lipid Metabolism. PLOS ONE 2013, 8, e58856.
- Galmes, R.; Houcine, A.; Van Vliet, A.; Agostinis, P.; Jackson, C.L.; Giordano, F. ORP5/ORP8 localize to endoplasmic reticulum–mitochondria contacts and are involved in mitochondrial function. EMBO Rep. 2016, 17, 800–810.
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324.
- Massey, J.B. Membrane and protein interactions of oxysterols. Curr. Opin. Lipidol. 2006, 17, 296–301.
- Chang, T.-Y.; Chang, C.C.; Ohgami, N.; Yamauchi, Y. Cholesterol Sensing, Trafficking, and Esterification. Annu. Rev. Cell Dev. Boil. 2006, 22, 129–157.
- Beh, C.T.; Rine, J. A role for yeast oxysterol-binding protein homologs in endocytosis and in the maintenance of intracellular sterol-lipid distribution. J. Cell Sci. 2004, 117, 2983–2996.
- Maeda, K.; Anand, K.; Chiapparino, A.; Kumar, A.; Poletto, M.; Kaksonen, M.; Gavin, A.-C. Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature 2013, 501, 257–261.
- Wang, P.; Zhang, Y.; Li, H.; Chieu, H.K.; Munn, A.L.; Yang, H. AAA ATPases regulate membrane association of yeast oxysterol binding proteins and sterol metabolism. EMBO J. 2005, 24, 2989–2999.
- Wang, P.; Duan, W.; Munn, A.L.; Yang, H. Molecular characterization of Osh6p, an oxysterol binding protein homolog in the yeast Saccharomyces cerevisiae. FEBS J. 2005, 272, 4703–4715.
- Suchanek, M.; Hynynen, R.; Wohlfahrt, G.; Lehto, M.; Johansson, M.; Saarinen, H.; Radzikowska, A.; Thiele, C.; Olkkonen, V.M. The mammalian oxysterol-binding protein-related proteins (ORPs) bind 25-hydroxycholesterol in an evolutionarily conserved pocket. Biochem. J. 2007, 405, 473–480.
- Zhou, T.; Li, S.; Zhong, W.; Vihervaara, T.; Béaslas, O.; Perttilä, J.; Luo, W.; Jiang, Y.; Lehto, M.; Olkkonen, V.M.; et al. OSBP-Related Protein 8 (ORP8) Regulates Plasma and Liver Tissue Lipid Levels and Interacts with the Nucleoporin Nup62. PLOS ONE 2011, 6, e21078.
- Van Kampen, E.; Béaslas, O.; Hildebrand, R.B.; Lammers, B.; Van Berkel, T.J.C.; Olkkonen, V.M.; Van Eck, M. Orp8 Deficiency in Bone Marrow-Derived Cells Reduces Atherosclerotic Lesion Progression in LDL Receptor Knockout Mice. PLOS ONE 2014, 9, e109024.
- Vihervaara, T.; Käkelä, R.; Liebisch, G.; Tarasov, K.; Schmitz, G.; Olkkonen, V.M. Modification of the lipidome in RAW264.7 macrophage subjected to stable silencing of oxysterol-binding proteins. Biochimie 2013, 95, 538–547.
- Li, B.; Fan, J.; Chena, N. A Novel Regulator of Type II Diabetes: MicroRNA-143. Trends Endocrinol. Metab. 2018, 29, 380–388.
- Blumensatt, M.; Greulich, S.; De Wiza, D.H.; Mueller, H.; Maxhera, B.; Rabelink, M.J.; Hoeben, R.C.; Akhyari, P.; Al-Hasani, H.; Ruige, J.B.; et al. Activin A impairs insulin action in cardiomyocytes via up-regulation of miR-143. Cardiovasc. Res. 2013, 100, 201–210.
- Blumensatt, M.; Wronkowitz, N.; Wiza, C.; Cramer, A.; Mueller, H.; Rabelink, M.J.; Hoeben, R.C.; Eckel, J.; Sell, H.; Ouwens, D.M. Adipocyte-derived factors impair insulin signaling in differentiated human vascular smooth muscle cells via the upregulation of miR-143. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2014, 1842, 275–283.
- Zhong, W.; Zhou, Y.; Li, J.; Mysore, R.; Luo, W.; Li, S.; Chang, M.-S.; Olkkonen, V.M.; Yan, D. OSBP-related protein 8 (ORP8) interacts with Homo sapiens sperm associated antigen 5 (SPAG5) and mediates oxysterol interference of HepG2 cell cycle. Exp. Cell Res. 2014, 322, 227–235.
- Li, J.; Zheng, X.; Lou, N.; Zhong, W.; Yan, D. Oxysterol binding protein-related protein 8 mediates the cytotoxicity of 25-hydroxycholesterol. J. Lipid Res. 2016, 57, 1845–1853.
- Guo, X.; Zhang, L.; Fan, Y.; Zhang, D.; Qin, L.; Dong, S.; Li, G. Oxysterol-Binding Protein-Related Protein 8 Inhibits Gastric Cancer Growth Through Induction of ER Stress, Inhibition of Wnt Signaling, and Activation of Apoptosis. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2017, 25, 799–808.
- Zhang, Z.; Gao, W.; Zhou, L.; Chen, Y.; Qin, S.; Zhang, L.; Liu, J.; He, Y.; Lei, Y.; Chen, H.-N.; et al. Repurposing Brigatinib for the Treatment of Colorectal Cancer Based on Inhibition of ER-phagy. Theranostics 2019, 9, 4878–4892.
- Ishikawa, S.; Nagai, Y.; Masuda, T.; Koga, Y.; Nakamura, T.; Imamura, Y.; Takamori, H.; Hirota, M.; Funakosi, A.; Fukushima, M.; et al. The role of oxysterol binding protein-related protein 5 in pancreatic cancer. Cancer Sci. 2009, 101, 898–905.
- Lippincott-Schwartz, J.; Phair, R. Lipids and cholesterol as regulators of traffic in the endomembrane system. Annu. Rev. Biophys. 2010, 39, 559–578.
- Wang, Y.; Thiele, C.; Huttner, W.B. Cholesterol is Required for the Formation of Regulated and Constitutive Secretory Vesicles from the trans-Golgi Network. Traffic 2000, 1, 952–962.
- Runz, H.; Miura, K.; Weiss, M.; Pepperkok, R. Sterols regulate ER-export dynamics of secretory cargo protein ts-O45-G. EMBO J. 2006, 25, 2953–2965.
- Ridsdale, A.; Denis, M.; Gougeon, P.-Y.; Ngsee, J.K.; Presley, J.F.; Zha, X. Cholesterol Is Required for Efficient Endoplasmic Reticulum-to-Golgi Transport of Secretory Membrane Proteins. Mol. Boil. Cell 2006, 17, 1593–1605.
- Nabi, I.R.; Le, P.U. Caveolae/raft-dependent endocytosis. J. Cell Boil. 2003, 161, 673–677.
- Kozminski, K.G.; Alfaro, G.; Dighe, S.; Beh, C.T. Homologues of Oxysterol-Binding Proteins Affect Cdc42p- and Rho1p-Mediated Cell Polarization in Saccharomyces cerevisiae. Traffic 2006, 7, 1224–1242.
- Béaslas, O.; Vihervaara, T.; Li, J.; Laurila, P.-P.; Yan, D.; Olkkonen, V.M. Silencing of OSBP-related protein 8 (ORP8) modifies the macrophage transcriptome, nucleoporin p62 distribution, and migration capacity. Exp. Cell Res. 2012, 318, 1933–1945.




