Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Moo-Seung Lee | + 1975 word(s) | 1975 | 2021-06-16 07:36:46 | | | |
| 2 | Vicky Zhou | Meta information modification | 1975 | 2021-06-23 11:10:29 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Lee, M. Escherichia coli Shiga Toxins. Encyclopedia. Available online: https://encyclopedia.pub/entry/11187 (accessed on 07 February 2026).
Lee M. Escherichia coli Shiga Toxins. Encyclopedia. Available at: https://encyclopedia.pub/entry/11187. Accessed February 07, 2026.
Lee, Moo-Seung. "Escherichia coli Shiga Toxins" Encyclopedia, https://encyclopedia.pub/entry/11187 (accessed February 07, 2026).
Lee, M. (2021, June 23). Escherichia coli Shiga Toxins. In Encyclopedia. https://encyclopedia.pub/entry/11187
Lee, Moo-Seung. "Escherichia coli Shiga Toxins." Encyclopedia. Web. 23 June, 2021.
Copy Citation
Escherichia coli (EHEC) and Shigella dysenteriae serotype 1 are enterohemorrhagic bacteria that induce hemorrhagic colitis. This, in turn, may result in potentially lethal complications, such as hemolytic uremic syndrome (HUS), which is characterized by thrombocytopenia, acute renal failure, and neurological abnormalities. Both species of bacteria produce Shiga toxins (Stxs), a phage-encoded exotoxin inhibiting protein synthesis in host cells that are primarily responsible for bacterial virulence.
Shiga toxins
Shiga toxin types 1 and 2
Shiga-toxin-producing (STEC)
commensal microbes
bacterial toxins
gut microbiota
hemolytic uremic syndrome (HUS)
1. Introduction
Numerous supportive public health measures have led people to erroneously believe that epidemics of many bacterial infectious diseases are no longer a serious health risk. However, Shiga-toxin-producing Escherichia coli (STEC) still poses a threat to public health. Shiga toxin (Stx) is prototypically synthesized by the bacterium Shigella dysenteriae serotype 1, with genetically and structurally related toxin variants produced by certain serotypes of E. coli, including enterohaemorrhagic strains of E. coli (EHEC) [1]. Bacillary dysentery due to infection by Stx-producing bacteria, characterized by acute infectious diarrhea, primarily affects children aged <5 years [2]. Endemic bacillary dysentery occurs globally, including in portions of Africa, Southeast Asia, and the Indian subcontinent, with an estimated 2–7 per 1000 children per year requiring clinical care and 164,300 deaths per year attributable to shigellosis [3]. By contrast, STEC-associated illnesses in young children are more prevalent in developed countries, in which residents consume higher levels of beef and beef products [4][5]. Major outbreaks of diarrheal diseases caused by EHEC may be due to the ingestion of foods such as uncooked meat, unpasteurized milk, water contaminated with these bacteria, and by the contamination of foods used in the preparation of fast-food [6]. Perhaps the largest outbreak of hemorrhagic colitis was caused by an O157 infection in and around Sakai City, Japan, in 1996, which resulted in approximately 1000 hospitalizations among 7000 infected cases [7]. Many non-O157 STEC serotypes have been increasingly reported, and a massive outbreak caused by the hybrid STEC/enteroaggregative E. coli (EAEC) O104:H4 strain occurred in northern Germany from May to June 2011 [8]. More recently, 439 outbreaks and 5 deaths caused by EHEC-contaminated romaine lettuce were reported in multiple states of the United States in 2018 [9].
Stx-producing bacteria have received substantial attention as emergent pathogens due to the dangerous toxins they produce. These exotoxins are the principal virulence factors associated with the pathogenesis of bloody diarrheal diseases, bacillary dysentery, and hemorrhagic colitis progressing to acute renal failure in infected patients, primarily in children. This phenomenon, collectively referred to as hemolytic uremic syndrome (HUS), is the leading cause of pediatric acute renal failure in many countries, including countries in the European Union and the United States [10][11]. Both Stxs and the inflammatory innate immune cells activated by these toxins contribute to the pathogenesis of HUS by rendering blood vessels in the colon, kidney, and central nervous system (CNS) more sensitive to the detrimental action of Stxs. Studies in animals found that treatment with purified Stxs induces intestinal and renal epithelial and endothelial cells to express neutrophil and monocyte chemoattractants. This, in turn, induces the infiltration of peripheral blood mononuclear cells (PBMCs) into the lamina propria and kidneys [12][13]. These findings suggest that the infiltration of inflammatory cells into sites of toxin-induced damage causes the localized expression of cytokines, which in turn facilitate vascular damage via immunopathological reactions.
Studies on Stxs-induced host signaling pathways have indicated that these toxins, which act as multifunctional bacterial proteins, promote ribotoxic stress, apoptosis, endoplasmic reticulum (ER) stress, inflammatory responses, and autophagy in host cells [14]. In addition to the toxigenic and immunopathological potential of Stxs in patients, these toxins interact with multiple cell types in vitro and are responsible for the pathogenic characteristics of HUS in animal models. Although the Stxs-mediated pathogenesis of HUS is not fully understood, comprehensive knowledge of the role of Stxs in altering the composition (also referred to as ‘dysbiosis’) of the intestinal microbiota in a host infected with EHEC compared with a healthy control must be used to help identify the host factors or the commensal microbial-derived products that exacerbate tissue damage or protect against intoxication caused by toxin-producing bacteria. Studies have evaluated the precise correlations between Stx-mediated pathogenesis and intestinal indigenous commensal microbes.
2. Effects of Probiotics on STEC and Stxs in the Gut
E. coli is mainly found in the intestinal cecum and colon of mammals and resides in the mucosal layer, from which it moves into the intestinal lumen and is excreted into the feces. Pathogenic E. coli-mediated diseases, such as food poisoning, intestinal tissue damage, and particularly bloody diarrhea in young children, are major concerns in public health and medical expense-related economic problems worldwide. More importantly, certain strains of STEC in the gut may cause severe extraintestinal or extrarenal illnesses in humans. Certain Gb3-expressing cell types in the gut, such as Paneth cells, may serve as portals for ingress of Stxs. Several epidemiological studies have shown that infection with STEC isolates expressing Stx2a is more pathogenic than infection with strains producing Stx1a or Stx1a+Stx2a [4][15][16]. Infection with strains producing Stx2a may lead to extraintestinal complications, although the cause of the latter is likely to be multi-factorial and can include the constitutive regulation of stx gene expression; antibiotic usage during the prodromal diarrheal phase, inducing the phage-mediated lytic cycle; the presence or absence of additional E. coli virulence factors; and variations in host responses to toxins, which all contribute to the outcome of infection. Moreover, stx genes are carried in the genomes of temperate phages [17][18], located in the late gene region downstream of the late promoters and upstream of the lysis cassette, highly expressed upon activation of phage-mediated lytic cycle and the toxin subunits assembled in the periplasm are secreted by the lytic cycle [19]. Therefore, antibiotics are not recommended for patients with HUS because they activate the phage-mediated lytic cycle in STEC [20] that lyses bacterial host cells to release Stxs and free phage particles that can infect other bacteria and transduce stx genes [21][22][23][24][25][26]. Given that antibiotic therapy is not indicated for these reasons, besides numerous therapeutic approaches utilizing Stxs-specific neutralizing antibodies, toxin receptor analogs, and vaccinations [27], studies have attempted to increase the population of intestinal microbiota that can inhibit STEC colonization and/or Stx expression (Table 1, Figure 1), thereby limiting HUS development.
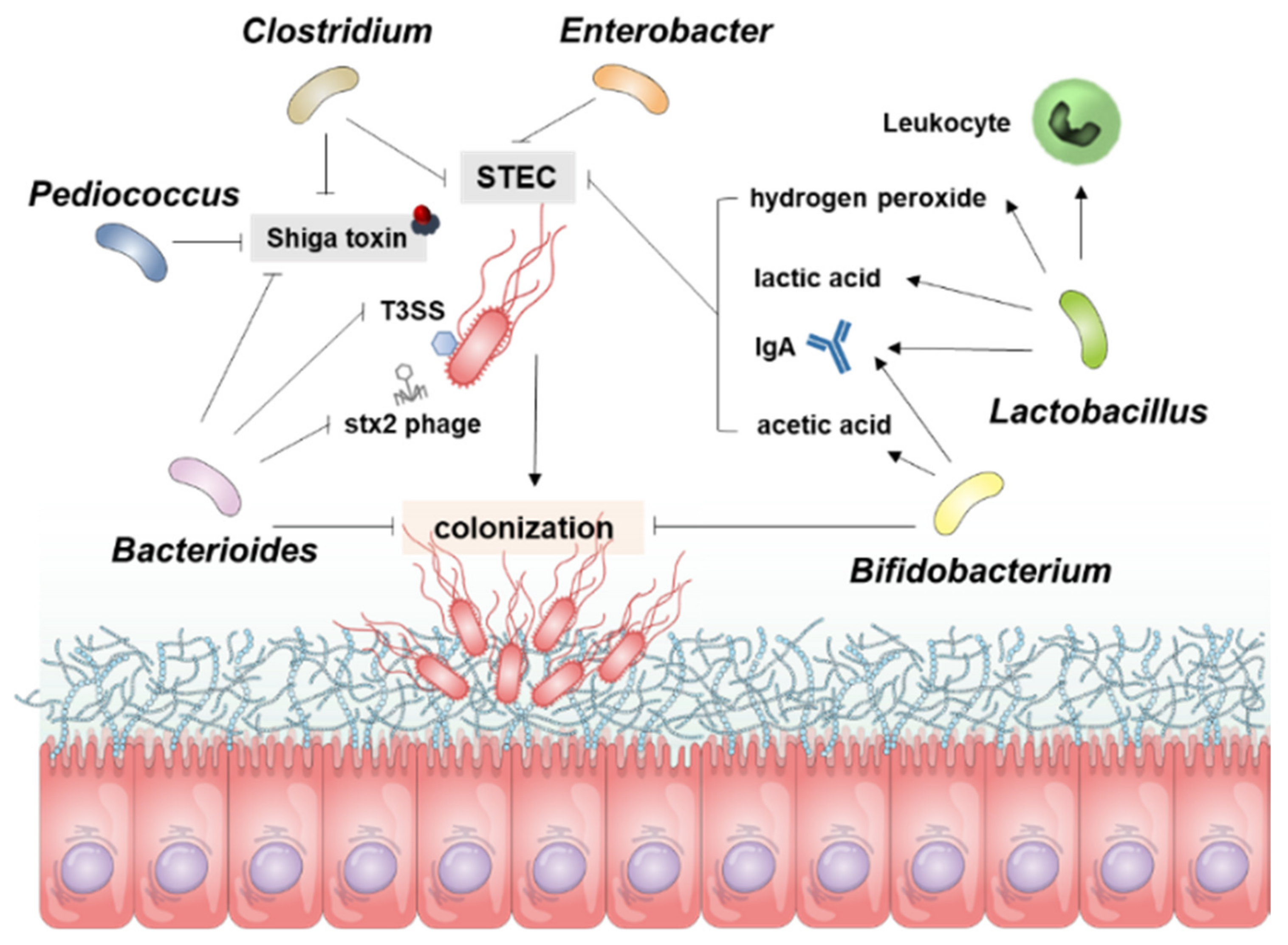 Figure 1. Overview for the Resistance Mechanism of gut microbiota to STEC Infection. The resistance of microbial guns to control STEC works in several ways, and the diagram shows the role of each bacterium in STEC. Bacterioides inhibit Stx production, control direct bacteria and inhibit the colonization of STEC. Bifidobacterium also inhibits STEC colonization and controls STEC proliferation through acetic acid and IgA. Lactobacillus is involved in inhibiting STEC proliferation through the production of hydrogen peroxide, lactic acid, IgA, and leukocyte activity. Pediococcus, Clostridium, and Enterobacter are involved in inhibiting STEC proliferation or controlling Stx production.
Figure 1. Overview for the Resistance Mechanism of gut microbiota to STEC Infection. The resistance of microbial guns to control STEC works in several ways, and the diagram shows the role of each bacterium in STEC. Bacterioides inhibit Stx production, control direct bacteria and inhibit the colonization of STEC. Bifidobacterium also inhibits STEC colonization and controls STEC proliferation through acetic acid and IgA. Lactobacillus is involved in inhibiting STEC proliferation through the production of hydrogen peroxide, lactic acid, IgA, and leukocyte activity. Pediococcus, Clostridium, and Enterobacter are involved in inhibiting STEC proliferation or controlling Stx production.Table 1. Abilities of probiotics to regulate Stx production and STEC virulence.
| Effect | Activity | Mediator | Model | Intestine In-/Out-Side |
Genus | Species | Ref |
|---|---|---|---|---|---|---|---|
| Inhibition | Inhibit growth of E. coli O157:H7 | Butyric, lactic acid | Gnotobiotic mice | Inside | Clostridium | butyricum | [28] |
| N/D | Gnotobiotic mice | Inside | Bacteroides | fragilis | [29] | ||
| N/D | Gnotobiotic mice | Inside | Lactobacillus | reuteri | [30] | ||
| N/D | Cattle | Inside | Lactobacillus | acidophilus | [31] | ||
| N-acetylglucosamine (NAG) and N-acetylneuraminic acid (NANA) | BALB/c mice Bovine rumen |
Inside | Bacteroides | thetaiotaomicron | [32] | ||
| Reuterin | fluid | Inside | Lactobacillus | reuteri | [33] | ||
| Reduce autoinducer-2 (AI-2) production | E. coli O157:H7 | N/D | Lactobacillus | acidophilus | [34] | ||
| Hydrogen peroxide | Raw chicken meat | N/D | Lactobacillus | lactis | [35] | ||
| Lactic acid | E. coli O157:H7 | N/D | Lactobacillus | casei | [36] | ||
| Decrease pH | E. coli O157:H7 | N/D | Bifidobacterium | bifidum | [37] | ||
| Acetic acid | BALB/c mice | Inside | Bifidobacterium | breve | [38] | ||
| Nutrition competition (carbon, nitrogen) |
Lettuce | N/D | Enterobacter | asburiae | [39] | ||
| Production of anti-Stx1 and -Stx2 IgA in the colon | Infant rabbits | Inside | Lactobacillus | casei | [40] | ||
| IgA | BALB/c mice | Inside | Bifidobacterium | thermacidophilum | [41] | ||
| Regulate host immunity | Upregulate intestinal anti-E. coli IgA responses | BALB/c and C57BL/6 mice |
Inside | Lactobacillus | rhamnosus | [42] | |
| Blood leukocyte activity | |||||||
| Inhibit translocation of E. coli O157:H7 | |||||||
| Production of anti-Stx1 and -Stx2 IgA in the colon | Infant rabbits | Inside | Lactobacillus | casei | [40] | ||
| Increase phagocytic activity | BALB/c and C57BL/6 mice | Outside | Bifidobacterium | lactis | [43] | ||
| Increase production of IgA against E. coli O157:H7 | BALB/c and C57BL/6 mice | Inside | Bifidobacterium | lactis | [43] | ||
| Increase production of IgG and IgM against E. coli O157:H7 | BALB/c mice | Outside | Bifidobacterium | thermacidophilum | [41] | ||
| IgA | BALB/c mice | Inside | Bifidobacterium | thermacidophilum | [41] | ||
| Reduce Stx production | N/D | Gnotobiotic mice | Inside | Bifidobacterium | infantis | [44] | |
| N/D | Gnotobiotic mice | Inside | Bifidobacterium | longum | [44] | ||
| Butyric, lactic acid | Gnotobiotic mice | Inside | Clostridium | butyricum | [28] | ||
| N/D | Gnotobiotic mice | Inside | Bacteroides | fragilis | [29] | ||
| Acetic acid | BALB/c mice | Inside | Bifidobacterium | breve | [38] | ||
| Production of anti-Stx1 and -Stx2 IgA in the colon | Infant rabbits | Inside | Lactobacillus | casei | [40] | ||
| N/D | E. coli O157:H7 | N/D | Bacteroides | thetaiotaomicron | [45] | ||
| Uptake vitamin B12 | E. coli O157:H7 | N/D | Bacteroides | thetaiotaomicron | [46] | ||
| Suppress kidney necrosis induced by E. coli O157:H7 | N/D | Gnotobiotic mice | Inside | Lactobacillus | reuteri | [30] | |
| Repress T3SS of E. coli O157:H7 | NANA and NAG | E. coli O157:H7 | N/D | Bacteroides | thetaiotaomicron | [32] | |
| Reduce intestinal injuries after E. coli O157:H7 infection | Production of anti-Stx1 and -Stx2 IgA in the colon | Infant rabbits | Inside | Lactobacillus | casei | [40] | |
| Increased production of IgG and IgM against E. coli O157:H7 | BALB/c mice | Inside | Bifidobacterium | thermacidophilum | [41] | ||
| Inhibit stx2 phage particle release | N/D | E. coli O157:H7 | N/D | Bacteroides | thetaiotaomicron | [45] | |
| Reduce Stx2 gene expression | Inhibition of phage production | E. coli O153:H25 | N/D | Bacteroides | thetaiotaomicron | [47] | |
| Organic acid produced by probiotics | E. coli O157:H7 | N/D | Pediococcus | pentosaceus | [48] | ||
| Lactobacillus | rhamnosus GG | ||||||
| Bifidobacterium | thermophilum | ||||||
| Reduce attaching and effacing lesions of E. coli O157:H7 | Spent medium | ICR mice | Inside | Lactobacillus | acidophilus | [49] | |
| HeLa cells | N/D | ||||||
| Hep-2 cells | |||||||
| S-layer protein | Hep-2 cells | N/D | Lactobacillus | helveticus | [50] | ||
| T84 cells | |||||||
| N/D | Hep-2 cells | N/D | Lactobacillus | rhamnosus | [51] | ||
| T84 cells | |||||||
| Enhancement | Increase toxin receptor expression on host cells | Butyrate | E. coli O157:H7 | N/D | N/D | N/D | [52] |
| Enhance toxin production | Bacteriophage transfer | CD-1 mice | Inside | Escherichia | coli | [53] | |
| [54] | |||||||
| Damage of E. coli O157:H7 DNA using DNase colicins | E. coli O157:H7 | N/D | Escherichia | coli | [55] | ||
| Increase the expression of the virulence genes of E. coli O157:H7 | Regulate Cra, a transcription factor for virulence genes of E. coli O157:H7 | C3H/HeJ mice | Inside | Bacteroides | thetaiotaomicron | [56] | |
| Exacerbate weight loss after infection | E. coli O157:H7 | Inside | Bacteroides | thetaiotaomicron | [56] | ||
| Enhance colonization | Butyrate | E. coli O157:H7 | N/D | N/D | N/D | [52] | |
| Secrete proteases that cleave the translocon of the T3SS | E. coli O157:H7 | N/D | Bacteroides | thetaiotaomicron | [57] | ||
| Fucose | E. coli O157:H7 | N/D | Bacteroides | thetaiotaomicron | [58] | ||
| Enhance T3SS of E. coli O157:H7 | Secrete proteases that cleave the translocon of the T3SS | E. coli O157:H7 | N/D | Bacteroides | thetaiotaomicron | [57] | |
| Increase E. coli O157:H7 motility | 4-Methyl benzoic acid | E. coli O157:H7 | N/D | N/D | N/D | [59] | |
| 3,4-Dimethylbenzoic acid | |||||||
| Hexanoic acid | |||||||
| Heptanoic acid |
3. Conclusions and Future Perspectives
Attempts to identify Stxs-induced risk factors in host cellular responses have revealed that these toxins have a wide range of novel properties that are associated with pathogenesis. Although studies have described Stx-induced signaling pathways that are associated with tissue damage, inflammation, and complement activation resulting from the immunopathological responses to these bacterial toxins, many details on the interfaces between STEC and commensal microbiota (Figure 1) remain to be determined. In particular, no coherent mechanism to date has defined the targets for intervention in HUS disease progression that might explain the dynamic immune modulation, or the mediators of inflammation associated with Stxs interacting with commensal gut microbiota. Additional studies are needed to better understand the intricate pathophysiology involving Stxs-associated bacterial communities in the gut. The precise mechanism by which changes of intestinal microbiota in response to EHEC Stxs may enhance host defense or exacerbate multi-organ damage warrants further investigation. These studies may help to identify potential targets for the disruption of innate immune responses or the protection of primary organs from damage induced by Stx1a and Stx2a.
References
- Kaper, J.B.; O’Brien, A.D. Overview and Historical Perspectives. Microbiol Spectr. 2014, 2.
- Parisot, M.; Parez, N.; Boukhari, R.; Breurec, S.; Jolivet, A. Shigella infection in children under 5 years old in western French Guiana. Epidemiol. Infect. 2018, 146, 980–984.
- Kotloff, K.L.; Riddle, M.S.; Platts-Mills, J.A.; Pavlinac, P.; Zaidi, A.K.M. Shigellosis. Lancet 2018, 391, 801–812.
- Griffin, P.M.; Tauxe, R.V. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 1991, 13, 60–98.
- Majowicz, S.E.; Scallan, E.; Jones-Bitton, A.; Sargeant, J.M.; Stapleton, J.; Angulo, F.J.; Yeung, D.H.; Kirk, M.D. Global incidence of human Shiga toxin-producing Escherichia coli infections and deaths: A systematic review and knowledge synthesis. Foodborne Pathog. Dis. 2014, 11, 447–455.
- Heredia, N.; García, S. Animals as sources of food-borne pathogens: A review. Anim. Nutr. 2018, 4, 250–255.
- Michino, H.; Araki, K.; Minami, S.; Takaya, S.; Sakai, N.; Miyazaki, M.; Ono, A.; Yanagawa, H. Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am. J. Epidemiol. 1999, 150, 787–796.
- Rasko, D.A.; Webster, D.R.; Sahl, J.W.; Bashir, A.; Boisen, N.; Scheutz, F.; Paxinos, E.E.; Sebra, R.; Chin, C.S.; Iliopoulos, D.; et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 2011, 365, 709–717.
- Centers for Disease Control and Prevention (CDC). Available online: (accessed on 1 April 2021).
- Scallan, E.; Mahon, B.E.; Hoekstra, R.M.; Griffin, P.M. Estimates of illnesses, hospitalizations and deaths caused by major bacterial enteric pathogens in young children in the United States. Pediatr. Infect. Dis. J. 2013, 32, 217–221.
- European Food Safety Authority; The European Centre for Disease Prevention and Control. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, e05500.
- Lee, M.S.; Kwon, H.; Nguyen, L.T.; Lee, E.Y.; Lee, C.Y.; Choi, S.H.; Kim, M.H. Shiga Toxins Trigger the Secretion of Lysyl-tRNA Synthetase to Enhance Proinflammatory Responses. J. Microbiol. Biotechnol. 2016, 26, 432–439.
- Jeong, Y.J.; Park, S.K.; Yoon, S.J.; Park, Y.J.; Lee, M.S. Experimental in vivo models of bacterial Shiga toxin-associated hemolytic uremic syndrome. J. Microbiol. Biotechnol. 2018.
- Lee, M.S.; Koo, S.; Jeong, D.G.; Tesh, V.L. Shiga Toxins as Multi-Functional Proteins: Induction of Host Cellular Stress Responses, Role in Pathogenesis and Therapeutic Applications. Toxins 2016, 8, 77.
- Melton-Celsa, A.R. Shiga Toxin (Stx) Classification, Structure, and Function. Microbiol. Spectr. 2014, 2, 37–53.
- FAO; WHO STEC Expert Group. Hazard Identification and Characterization: Criteria for Categorizing Shiga Toxin-Producing Escherichia coli on a Risk Basis(dagger). J. Food Prot. 2019, 82, 7–21.
- Herold, S.; Karch, H.; Schmidt, H. Shiga toxin-encoding bacteriophages--genomes in motion. Int. J. Med. Microbiol. 2004, 294, 115–121.
- O’Brien, A.D.; Newland, J.W.; Miller, S.F.; Holmes, R.K.; Smith, H.W.; Formal, S.B. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 1984, 226, 694–696.
- Wagner, P.L.; Livny, J.; Neely, M.N.; Acheson, D.W.; Friedman, D.I.; Waldor, M.K. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol. Microbiol. 2002, 44, 957–970.
- Tarr, P.I.; Gordon, C.A.; Chandler, W.L. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 2005, 365, 1073–1086.
- Acheson, D.W.; Reidl, J.; Zhang, X.; Keusch, G.T.; Mekalanos, J.J.; Waldor, M.K. In vivo transduction with shiga toxin 1-encoding phage. Infect. Immun. 1998, 66, 4496–4498.
- James, C.E.; Stanley, K.N.; Allison, H.E.; Flint, H.J.; Stewart, C.S.; Sharp, R.J.; Saunders, J.R.; McCarthy, A.J. Lytic and lysogenic infection of diverse Escherichia coli and Shigella strains with a verocytotoxigenic bacteriophage. Appl. Environ. Microbiol. 2001, 67, 4335–4337.
- Kratochvil, S. [Clinical experiences with hypnosis in the Czech republic (II)]. Psychiatr. Neurol. Med. Psychol. Beih 1981, 28, 46–51.
- Muniesa, M.; Blanco, J.E.; de Simon, M.; Serra-Moreno, R.; Blanch, A.R.; Jofre, J. Diversity of stx2 converting bacteriophages induced from Shiga-toxin-producing Escherichia coli strains isolated from cattle. Microbiology 2004, 150, 2959–2971.
- Schmidt, H.; Bielaszewska, M.; Karch, H. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage phi3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 1999, 65, 3855–3861.
- Tóth, I.; Schmidt, H.; Dow, M.; Malik, A.; Oswald, E.; Nagy, B. Transduction of porcine enteropathogenic Escherichia coli with a derivative of a shiga toxin 2-encoding bacteriophage in a porcine ligated ileal loop system. Appl. Environ. Microbiol. 2003, 69, 7242–7247.
- Mühlen, S.; Dersch, P. Treatment Strategies for Infections With Shiga Toxin-Producing Escherichia coli. Front. Cell Infect. Microbiol. 2020, 10, 169.
- Takahashi, M.; Taguchi, H.; Yamaguchi, H.; Osaki, T.; Komatsu, A.; Kamiya, S. The effect of probiotic treatment with Clostridium butyricum on enterohemorrhagic Escherichia coli O157:H7 infection in mice. FEMS Immunol. Med. Microbiol. 2004, 41, 219–226.
- Saito, K.; Suzuki, R.; Koyanagi, Y.; Isogai, H.; Yoneyama, H.; Isogai, E. Inhibition of enterohemorrhagic Escherichia coli O157:H7 infection in a gnotobiotic mouse model with pre-colonization by Bacteroides strains. Biomed. Rep. 2019, 10, 175–182.
- Eaton, K.A.; Honkala, A.; Auchtung, T.A.; Britton, R.A. Probiotic Lactobacillus reuteri ameliorates disease due to enterohemorrhagic Escherichia coli in germfree mice. Infect. Immun. 2011, 79, 185–191.
- Peterson, R.E.; Klopfenstein, T.J.; Erickson, G.E.; Folmer, J.; Hinkley, S.; Moxley, R.A.; Smith, D.R. Effect of Lactobacillus acidophilus strain NP51 on Escherichia coil O157:H7 fecal shedding and finishing performance in beef feedlot cattle. J. Food Prot. 2007, 70, 287–291.
- Le Bihan, G.; Sicard, J.F.; Garneau, P.; Bernalier-Donadille, A.; Gobert, A.P.; Garrivier, A.; Martin, C.; Hay, A.G.; Beaudry, F.; Harel, J.; et al. The NAG Sensor NagC Regulates LEE Gene Expression and Contributes to Gut Colonization by Escherichia coli O157:H7. Front. Cell Infect. Microbiol. 2017, 7, 134.
- Bertin, Y.; Habouzit, C.; Duniere, L.; Laurier, M.; Durand, A.; Duchez, D.; Segura, A.; Thevenot-Sergentet, D.; Baruzzi, F.; Chaucheyras-Durand, F.; et al. Lactobacillus reuteri suppresses E. coli O157:H7 in bovine ruminal fluid: Toward a pre-slaughter strategy to improve food safety? PLoS ONE 2017, 12, e0187229.
- Medellin-Pena, M.J.; Wang, H.; Johnson, R.; Anand, S.; Griffiths, M.W. Probiotics affect virulence-related gene expression in Escherichia coli O157:H7. Appl. Environ. Microbiol. 2007, 73, 4259–4267.
- Brashears, M.M.; Reilly, S.S.; Gilliland, S.E. Antagonistic action of cells of Lactobacillus lactis toward Escherichia coli O157:H7 on refrigerated raw chicken meat. J. Food Prot. 1998, 61, 166–170.
- Ogawa, M.; Shimizu, K.; Nomoto, K.; Tanaka, R.; Hamabata, T.; Yamasaki, S.; Takeda, T.; Takeda, Y. Inhibition of in vitro growth of Shiga toxin-producing Escherichia coli O157:H7 by probiotic Lactobacillus strains due to production of lactic acid. Int. J. Food Microbiol. 2001, 68, 135–140.
- Massa, S.; Altieri, C.; Quaranta, V.; De Pace, R. Survival of Escherichia coli O157:H7 in yoghurt during preparation and storage at 4 degrees C. Lett. Appl. Microbiol. 1997, 24, 347–350.
- Asahara, T.; Shimizu, K.; Nomoto, K.; Hamabata, T.; Ozawa, A.; Takeda, Y. Probiotic bifidobacteria protect mice from lethal infection with Shiga toxin-producing Escherichia coli O157:H7. Infect. Immun. 2004, 72, 2240–2247.
- Cooley, M.B.; Chao, D.; Mandrell, R.E. Escherichia coli O157:H7 survival and growth on lettuce is altered by the presence of epiphytic bacteria. J. Food Prot. 2006, 69, 2329–2335.
- Ogawa, M.; Shimizu, K.; Nomoto, K.; Takahashi, M.; Watanuki, M.; Tanaka, R.; Tanaka, T.; Hamabata, T.; Yamasaki, S.; Takeda, Y. Protective effect of Lactobacillus casei strain Shirota on Shiga toxin-producing Escherichia coli O157:H7 infection in infant rabbits. Infect. Immun. 2001, 69, 1101–1108.
- Gagnon, M.; Kheadr, E.E.; Dabour, N.; Richard, D.; Fliss, I. Effect of Bifidobacterium thermacidophilum probiotic feeding on enterohemorrhagic Escherichia coli O157:H7 infection in BALB/c mice. Int. J. Food Microbiol. 2006, 111, 26–33.
- Shu, Q.; Gill, H.S. Immune protection mediated by the probiotic Lactobacillus rhamnosus HN001 (DR20) against Escherichia coli O157:H7 infection in mice. FEMS Immunol. Med. Microbiol. 2002, 34, 59–64.
- Shu, Q.; Gill, H.S. A dietary probiotic (Bifidobacterium lactis HN019) reduces the severity of Escherichia coli O157:H7 infection in mice. Med. Microbiol. Immunol. 2001, 189, 147–152.
- Yoshimura, K.; Matsui, T.; Itoh, K. Prevention of Escherichia coli O157:H7 infection in gnotobiotic mice associated with Bifidobacterium strains. Antonie Van Leeuwenhoek 2010, 97, 107–117.
- De Sablet, T.; Chassard, C.; Bernalier-Donadille, A.; Vareille, M.; Gobert, A.P.; Martin, C. Human microbiota-secreted factors inhibit shiga toxin synthesis by enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 2009, 77, 783–790.
- Cordonnier, C.; Le Bihan, G.; Emond-Rheault, J.G.; Garrivier, A.; Harel, J.; Jubelin, G. Vitamin B12 Uptake by the Gut Commensal Bacteria Bacteroides thetaiotaomicron Limits the Production of Shiga Toxin by Enterohemorrhagic Escherichia coli. Toxins 2016, 8, 14.
- Iversen, H.; Lindbäck, T.; L’Abee-Lund, T.M.; Roos, N.; Aspholm, M.; Stenfors Arnesen, L. The gut bacterium Bacteroides thetaiotaomicron influences the virulence potential of the enterohemorrhagic Escherichia coli O103:H25. PLoS ONE 2015, 10, e0118140.
- Carey, C.M.; Kostrzynska, M.; Ojha, S.; Thompson, S. The effect of probiotics and organic acids on Shiga-toxin 2 gene expression in enterohemorrhagic Escherichia coli O157:H7. J. Microbiol. Methods 2008, 73, 125–132.
- Medellin-Peña, M.J.; Griffiths, M.W. Effect of molecules secreted by Lactobacillus acidophilus strain La-5 on Escherichia coli O157:H7 colonization. Appl. Environ. Microbiol. 2009, 75, 1165–1172.
- Johnson-Henry, K.C.; Hagen, K.E.; Gordonpour, M.; Tompkins, T.A.; Sherman, P.M. Surface-layer protein extracts from Lactobacillus helveticus inhibit enterohaemorrhagic Escherichia coli O157:H7 adhesion to epithelial cells. Cell Microbiol. 2007, 9, 356–367.
- Sherman, P.M.; Johnson-Henry, K.C.; Yeung, H.P.; Ngo, P.S.; Goulet, J.; Tompkins, T.A. Probiotics reduce enterohemorrhagic Escherichia coli O157:H7- and enteropathogenic E. coli O127:H6-induced changes in polarized T84 epithelial cell monolayers by reducing bacterial adhesion and cytoskeletal rearrangements. Infect. Immun. 2005, 73, 5183–5188.
- Zumbrun, S.D.; Melton-Celsa, A.R.; Smith, M.A.; Gilbreath, J.J.; Merrell, D.S.; O’Brien, A.D. Dietary choice affects Shiga toxin-producing Escherichia coli (STEC) O157:H7 colonization and disease. Proc. Natl. Acad. Sci. USA 2013, 110, E2126–E2133.
- Gamage, S.D.; Strasser, J.E.; Chalk, C.L.; Weiss, A.A. Nonpathogenic Escherichia coli can contribute to the production of Shiga toxin. Infect. Immun. 2003, 71, 3107–3115.
- Gamage, S.D.; Patton, A.K.; Strasser, J.E.; Chalk, C.L.; Weiss, A.A. Commensal bacteria influence Escherichia coli O157:H7 persistence and Shiga toxin production in the mouse intestine. Infect. Immun. 2006, 74, 1977–1983.
- Toshima, H.; Yoshimura, A.; Arikawa, K.; Hidaka, A.; Ogasawara, J.; Hase, A.; Masaki, H.; Nishikawa, Y. Enhancement of Shiga toxin production in enterohemorrhagic Escherichia coli serotype O157:H7 by DNase colicins. Appl. Environ. Microbiol. 2007, 73, 7582–7588.
- Curtis, M.M.; Hu, Z.; Klimko, C.; Narayanan, S.; Deberardinis, R.; Sperandio, V. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe 2014, 16, 759–769.
- Cameron, E.A.; Curtis, M.M.; Kumar, A.; Dunny, G.M.; Sperandio, V. Microbiota and Pathogen Proteases Modulate Type III Secretion Activity in Enterohemorrhagic Escherichia coli. mBio 2018, 9.
- Pacheco, A.R.; Curtis, M.M.; Ritchie, J.M.; Munera, D.; Waldor, M.K.; Moreira, C.G.; Sperandio, V. Fucose sensing regulates bacterial intestinal colonization. Nature 2012, 492, 113–117.
- Tovaglieri, A.; Sontheimer-Phelps, A.; Geirnaert, A.; Prantil-Baun, R.; Camacho, D.M.; Chou, D.B.; Jalili-Firoozinezhad, S.; de Wouters, T.; Kasendra, M.; Super, M.; et al. Species-specific enhancement of enterohemorrhagic E. coli pathogenesis mediated by microbiome metabolites. Microbiome 2019, 7, 43.
More
Information
Subjects:
Microbiology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.8K
Revisions:
2 times
(View History)
Update Date:
23 Jun 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




