
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Kumar Sudesh | + 9464 word(s) | 9464 | 2021-06-21 05:15:23 | | | |
| 2 | Bruce Ren | -21 word(s) | 9443 | 2021-06-22 07:52:02 | | | | |
| 3 | Bruce Ren | Meta information modification | 9443 | 2021-06-22 11:22:44 | | | | |
| 4 | Bruce Ren | Meta information modification | 9443 | 2021-06-23 10:20:30 | | |
Video Upload Options
Rubber is an essential part of our daily lives with thousands of rubber-based products being made and used. Natural rubber undergoes chemical processes and structural modifications, while synthetic rubber, mainly synthetized from petroleum by-products are difficult to degrade safely and sustainably. The most prominent group of biological rubber degraders are Actinobacteria. Rubber degrading Actinobacteria contain rubber degrading genes or rubber oxygenase known as latex clearing protein (lcp). Rubber is a polymer consisting of isoprene, each containing one double bond. The degradation of rubber first takes place when lcp enzyme cleaves the isoprene double bond, breaking them down into the sole carbon and energy source to be utilized by the bacteria. Actinobacteria grow in diverse environments, and lcp gene containing strains have been detected from various sources including soil, water, human, animal, and plant samples.
1. Rubber—Polyisoprenes
1.1. Natural Rubber (NR)
A large fraction of the organic biomass on earth consists of biopolymers; such as polysaccharides, polyamino acids (proteins), polyconiferylalcohols (lignins), polyhydroxyalkanoic acids (PHAs), and polyisoprenes (rubbers) [1]. Natural rubbers (NR) farmed from Hevea brasiliensis Muell. Arg—comprising 99% of the world market—and Parthenium argentatum (guayule rubber) are produced commercially [2]. NR has a (cis)-1,4-polyisoprene as its polymer backbone. The backbone consists of isoprene units (C5H8) each containing one double bond in the cis or trans configuration, while 3 trans-isoprene units are found at one end of the molecule followed by several hundred to a few thousand cis-isoprene units (Figure 1) [3]. Most rubber-accumulating plants—and there are more than 2000 dicotyledons, and some fungi known to do so [4]—synthesize the polymer with the isoprene units in the cis-configuration [5]. Some species, such as Manilkara chicle or Palaquium gutta, however, synthesize the trans-polymer producing rubbers known as chicle or gutta-percha [6].
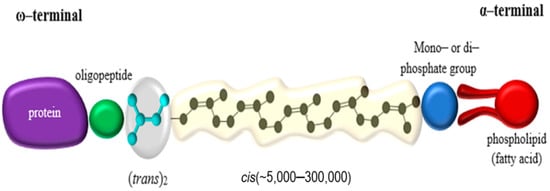
1.2. Production and Usage
1.3. Synthetic Rubber (IR)
1.4. Rubber Wastes—Mitigation and Drawbacks
2. Biodegradation: Roles of Microbes
2.1. Rubber Degraders: Who, Where, How?
2.1.1. Clear Zone Formers
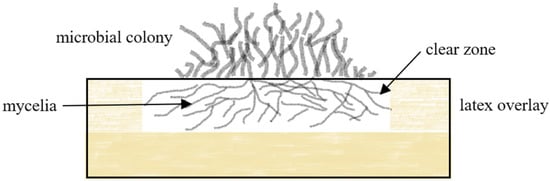
2.1.2. Adhesive Contact Strains
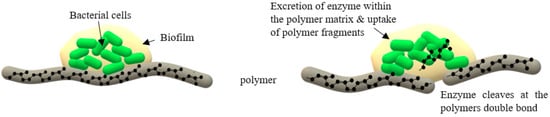
3. Rubber Degrading Enzymes: Rubber Oxygenase
| Lcp | RoxB | RoxA | |
|---|---|---|---|
| First Identified From | Streptomyces sp. strain K30 | Steroidobacter cummioxidans 35Y (Xanthomonas sp. 35Y) | |
| Accession no. | AY387589 | KY498024 | KC980911 |
| Metabolite Products | Tetra C20, oligoisoprene/ aldehyde and ketone terminal |
ODTDs | |
| Bacteria | Mostly Gram-positive | Gram-negative only | |
| Gene Length (bp) | ~1224–1227 | ~2037–2046 | |
| Size (kDa) | 43 | 73–74 | |
| Pathway | TAT | SEC | |
| Co-factor | b-type heme | c-type heme | |
| Mechanism to Cleavage Isoprene | Endo-type | Endo and Exo-type | |
| Major Metal Atoms in Protein Molecule | 1 Fe | 2 Fe | |
| Oxidation State of Iron | Fe3+ | Fe3+----O2 | |
3.1. Latex Clearing Protein (lcp)
3.1.1 Rubber Degradation by Lcp Enzyme
3.2. RoxA and RoxB
3.3. Others: Enzyme Mediator Systems
3.4. Unknown: Rubber Degradation in Fungi
4. Lcp Gene and Its Pathway
To identify whether the microbial rubber degradation is caused by lcp gene, one should amplify the gene and confirm the presence of conserve region. More complete sequencing would reveal lcp related genes, such as oxiAB genes, TAT secretion motif, and lcp regulator, which contribute to the rubber degradation pathway.
4.1. Conserved Region
Lcp from different species are related in amino acid sequence and share a common domain of unknown function DUF2236 together with other hypothetical proteins with different functional annotations [49][76]. DUF2236 sequences (1280 out of 6000) were analyzed and a 13-residue-long highly conserved region (KTRLVHAAVRHLL) in the primary amino acid sequence between K193 and L205 (in lcpK30) was identified [76].
Histidines are known to associate heme in proteins within this domain. Heme is an essential molecule and plays vital roles in many biological processes. It is evident that the three conserved DUF2236 residues (R164, T168, and H198) were highly conserved in the 495 lcp homologues by 98.8%, 99.6%, and 100%, respectively [76].
Three histidines in G. polyisoprenivorans lcp 1 (H195, 200 and 228) are highly conserved, and mineralization of rubber showed that histidine at position 195 is essential for strain VH2 to use rubber as sole source of carbon and energy [77]. It was further confirmed that heme-association is dependent on the presence of histidine at position 195 [77]. An analogue for H195 in lcpK30 is located at position 198, exchange of this amino acid (H198 or H195) led to the loss of heme [76].
4.2. Lcp Operon & Rubber Degradation Pathway
Although the mechanism of rubber degradation is not fully understood, several authors have postulated a rubber degrading pathway [41][56][78][79]. Here, we highlight the main sections of the putative rubber degradation pathway (Figure 4).
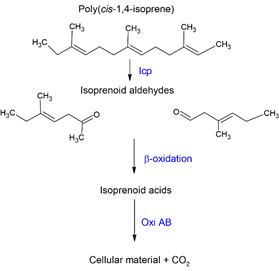
Figure 4. Schematic diagram representing the primary step of cis-1,4 isoprene biodegradation.
Three genes, namely lcp, oxiA, and oxiB, seem to exist as an operon having a polyisotronic mRNA. Lcp catalyzes cleavage of the C=C double bond of poly(cis-1,4-isoprene) found in NR and IR. In the process, oxygen is added across the double bonds, leading to a mixture of oligonucleotide-isoprenoids with terminal keto and aldehyde groups [80][81]. It is speculated that lcp binds and cleaves the polymer while oxiA and oxiB, located downstream of lcp, catabolize the degraded products [65]. The smaller oligomers are subsequently taken up by an Mce-protein (Mammalian cell entry) driven transport mechanism and are converted into the corresponding acids by a putative heterodimeric molybdenum hydroxylase, oxiAB, before entering the β-oxidation pathway [2][78]. OxiA is a small subunit critical for the iron-sulfur cluster center, while oxiB is a large subunit that possibly acts to bind molybdopterin cytosine dinucleotide cofactor [2]. OxiAB a molybdenum-dependent hydroxylase and is secreted extracellularly. The low molecular weight primary degradation products converted by oxidation to key compounds, including carboxyl groups, can then be taken up and metabolized by the cells via β-oxidation [54][39]. Other rubber degraders, such as Gordonia and Nocardia, do not possess an oxiAB homolog but contain aldehyde dehydrogenase genes (GPOL_c02580, GPOL_c37100) that theoretically perform a similar function [39].
4.3. Lcp Secretion Pathway
The lcp enzyme is exported by the (twin arginine translocation) Tat secretion pathway-dependent signal motif (RRxxLK) and a cleavage site for the signal sequence (AxA) was found [41]. In contrast to the Secretion (Sec) pathway which transports proteins in an unfolded manner (roxA and roxB), the Tat pathway serves to actively translocate folded proteins across a lipid membrane bilayer [82]. Folding secreted proteins before translocation would increase their stability and prevent protein aggregation intra- and extra-cellularly [2]. Tat-dependent proteins possess a variety of important functions that have an impact on bacterial cell physiology, such as respiratory energy metabolism, iron acquisition, stress response, and cell division [82].
4.4. Lcp Regulator
Two different types of TetR family of regulators (TFRs) were detected upstream of respective lcps for G. polyisoprenivorans VH2 (lcpVH2: dimer) and S. coelicolor lcpRBA3(2) (lcpRB: monomer). BLAST analyses revealed sequences coding for homologs of lcpRVH2 among other Actinobacteria, indicating a conserved regulation mechanism of lcps [77][83]. LcpRBA3(2) belonging, binds the upstream region of lcpA3(2) including the palindromic sequence (5′TATGTTAAT-N2-AAAATCACA-3′), to regulate the lcpA3(2) transcription, two DNA-binding HTH domains were found in the N-terminal of this protein [39]. Twelve strains that exhibited homolog gene coding for LcpRB and containing lcp gene were analyzed. Thereby, fifteen bases exhibited a conservation of at least 90%, including the inverted repeat that was detected in footprinting analyses inside the operator site of lcpRBA3(2) [83]. Notably, the lcpRB sequence was not always located directly next to lcp.
Recently, a novel key regulator in poly(cis-1,4-isoprene) degradation, cAMP receptor protein (CRPVH2), was detected for Gordonia polyisoprenivorans VH2, having 16 bp TGTGAN6TCACT motif [84]. CRP is also known as catabolite activator protein, CAP. CRPVH2 and G1xR has 79% sequence identity, both having a common oligomerization state, similar binding motifs and thus most 26 likely similar functions as global regulator [84]. G1xR is a well -studied global regulator of Corynebacterium glutamicum.
Groups of Lcp: Lcp protein characterization has been reported for G. polyisoprenivorans VH2, lcp1VH2 [79]; G. westfalica Kb1 [58], R. rhodochrous RPK1, lcpRPK1 [44]; Streptomyces sp. strain K30, lcpK30 [54]; Nocardia nova SH22a, lcpSH22a [85]; Nocardia sp. strain NVL3, lcpNVL3 [57]; N. farcinica E1, lcpE1 [86] and Solimonas fluminis HR-BB, LcpHR-B [59]. Phylogenetic analysis of lcp proteins showed that lcp gene might have evolved separately in each genus (Figure 5) [39]. Hence, the discovery and characterization of novel rubber degrading genera would be interesting and useful to decipher the distribution and characteristics of each lcp genes.
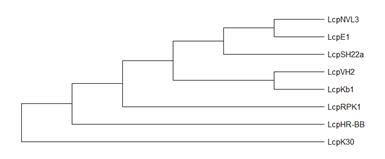
Figure 5. Phylogenetic tree of lcps using minimum evolution method was constructed using MEGA X [87][90]. Enzymes: lcpK30 (AAR25849); lcp1VH2 (ABV68923); lcpKb1 (ABV68924); lcpSH22a (WP025350295); (AMY60409); lcpNVL3 (API85527); lcpE1 (ABC59140) and lcpHR-BB (WP104231946).
5. Latex Clearing Protein: Synthesis
To further understand lcp activity and its implications for rubber degradation, we need to express and purify the enzyme. Large scale production of the active enzyme is needed for characterization studies and for industrial application of these enzymes in rubber waste treatment.
The synthesis of active lcp1VH2 (G. polyisoprenivorans VH2) in E. coli was first described by Hiessl et al., (2014). Purification of the protein was performed by immobilized metal affinity chromatography (IMAC) and size exclusion chromatography (SEC) [41]. The poor solubility of lcp1VH2 and the dramatic loss of target protein during the purification process, yielded low amounts of the desired protein. Following this, cultivation in Auto Induction Medium (AIM) and purification using ammonium sulphate precipitation yielded a 30-fold higher concentration of soluble lcp1VH2 (60 mg L−1). However, about 90% were inclusion bodies [88]. Using recombinant strains of E. coli, high yield production of soluble lcp1VH2 protein (223.5 mg L−1) through fed batch fermentations was successfully achieved [56]. Using a defined mineral salt medium [89] for E. coli cultivation, the group was able to significantly increase the biomass up to nearly 10-fold and the total lcp1VH2 yield up to 3.7-fold, reducing the costs for the medium by 75%.
RoxA was successfully expressed from S. cummioxidans 35Y with a yield of 15 mg L−1 of secreted protein. However, heterologous expression of roxA has not been successful to date .
6. Actinobacteria and Their Potential for Biodegrading Rubber
Actinobacteria is the largest group of bacterial rubber degraders identified today. In this section, we discuss the diversity and characteristics of Actinobacteria which make them favorable as rubber degraders. Many reports on the isolation and screening of rubber degrading bacteria have been published. However, with the advancement of genomic sequencing, we compiled a list of rubber degrading Actinobacteria which gives us an insight on the overall distribution of lcp genes and isolation source (see Section 6.2).
6.1. Diversity
Actinobacteria are one of the largest and most diverse phyla among the bacteria with a well described phylogeny. According to the latest classification, phylum Actinobacteria is divided into 6 classes, namely: Actinobacteria, Acidimicrobiia, Coriobacteriia, Nitriliruptoria, Rubrobacteria, and Thermoleophilia . The class Actinobacteria is further divided into 16 orders that are Actinomycetales, Actinopolysporales, Bifidobacteriales, Catenulisporales, Corynebacteriales, Frankiales, Glycomycetales, Jiangellales, Kineosporiales, Micrococcales, Micromonosporales, Propionibacteriales, Pseudonocardiales, Streptomycetales, Streptosporangiales, and Incertae sedis [90]. The genera of this class exhibit enormous diversity in terms of their morphology and physiology capabilities. Morphologies range from coccoid (e.g., Micrococcus) or rod-coccoid (e.g., Arthrobacter) to fragmenting hyphal forms (e.g., Nocardia sp.) or permanent and highly differentiated branched mycelium (e.g., Streptomyces sp.) [91]. Diverse metabolic properties, such as the production of extracellular enzymes and the formation of a wide variety of secondary metabolites, are also exhibited in this group .
6.2. Distribution
Actinobacteria members have adopted diverse lifestyles; they may be inhabitants of soil or aquatic environments (e.g., Streptomyces, Micromonospora, Rhodococcus, and Salinispora species), pathogens (e.g., Corynebacterium, Mycobacterium, Nocardia, Tropheryma, and Propionibacterium), solely soil inhabitants (Streptomyces), plant commensals (Leifsonia), plant symbionts (e.g., Frankia spp.), or gastrointestinal commensals (Bifidobacterium) [92]. The main microbial population is found in the surface layer of soil (106–109 cells gram−1), where they play a crucial role in recycling refractory biomaterials by decomposition and humus formation . Common genera in soil samples are Streptomyces (nearly 70%), Nocardia, and Micromonospora, although Actinoplanes and Streptosporangium are also generally encountered [93]. They live under the most diverse conditions, aerobic and anaerobic, at temperatures of 5–7 °C and 45–70 °C [94]. Thermophilic strains (e.g., Thermoactinomycetes, Streptomyces) are important for biotechnological investigations and the ability to grow at high temperatures makes them suitable candidates for rubber biodegradation. Thermostable enzymes are more resistant to industrial processes with high solvent or high salt concentrations and offer potentially faster reaction kinetics, and importantly, high temperatures in a fermentative process to prevent costly microbial contamination [95].
6.3. Genomic Characteristics of Actinobacteria
Actinobacteria have high G+C base content, ranging from 51% in some Corynebacteria to more than 70% in Streptomyces and Frankia [92]. However, the G+C content is less than 50% in Gardnerella vaginalis and Tropheryma whipplei. Of those analyzed, all Actinobacteria have a single chromosome, but the presence of large plasmids is not uncommon. Base composition is a fundamental property of genomes having a strong influence on gene function and regulation [96]. Bohlin et al. (2017) found that the coding regions in core genomes are significantly more GC-rich [97].
The genome content in bacteria is shaped by the interplay between vertical inheritance (clonal reproduction) with gene loss and acquisition, the latter either involving the genesis of new genes within a lineage or horizontal gene transfer (HGT) from other lineages [91]. Microbial genomes evolve dynamically by both losing and gaining genes, to create new species. HGT is held responsible for enhancing the competitiveness of bacteria in their natural environments [98]. Some Actinobacteria strains have a linear chromosome, whereas most of the genera harbor a circular chromosome, or undergo chromosomal circularization [99][100][101]. Actinobacteria with a more complex life cycle and structure, such as Nocardia, Actinoplanes, Micromonospora, Streptovericillium, Streptomyces, and Saccharopolyspora, generally have a linear chromosome [102][103][104].
In general, the chromosomes found in Actinobacteria are large, relative to other bacteria, with an average size of over 5 megabases (Mb) [98]. However, genome sizes vary widely: from less than 1 Mb in some Tropherma to over 12 Mb in some Streptomyces. The microbial genome or pan genome consists of a “core” section, representing about half the genome, and “accessory” elements including the ends of the linear structure . Essential genes lie within the core while genes whose products would be expected to be adaptive tend to be present in the arms [102][105]. Gene inactivation and loss are common in groups with a host-associated lifestyle; the host supplies many of the metabolic intermediates, thereby eliminating their need to maintain many biosynthetic genes [98]. Gross chromosomal rearrangements are ubiquitous among Streptomyces and affects nearly all life functions, e.g., differentiation, secondary metabolism, and response to environmental changes [106].
Plasmids (extrachromosomal DNA) can be divided into five main types: fertility F-plasmids, resistance plasmids, virulence plasmids, degradative plasmids, and finally Col plasmids [107]. They carry genes that usually confer beneficial properties and are required for the survival of the host. One of the unique features of Actinobacteria, especially the genus Streptomyces, is the presence of linear plasmids, sized from 12 to 600 kb, often termed mega-plasmids [108].
6.4. Latex Clearing Protein in the GenBank: Actinobacteria
The GenBank protein database recorded 248 Actinobacteria with a DUF2236 domain containing latex clearing protein (accessed April, 2020). These strains consist of 16 families, 29 genera and 134 species (Table 2). Most of the genes were from Streptomyces (20.6%), followed by Mycobacteriodes (17.4%), and Nocardia (11.7%). When we profile the 248 lcp genes using a phylogenetic approach, 12 distinct clusters were observed, suggesting that there may be 12 different groups of lcp genes (Supplementary Figure S1). By identifying the lcp groups, we could screen for lcp genes instead of using living cultures which are limited by screening method used. This is made easier to screen as most culture collection centres have started making DNA extract available upon request.
Within the groups, lcp genes seem to be closely related to the strain taxonomy, however, the genera can also be distributed among other clusters. Lcp of the same genera within the clusters could be due to gene duplication, or lcp genes from different genera within the same cluster could be contributed to gene transfer.
Jendrossek et al. (1997) screened 1220 Actinobacteria strains, where the majority of the rubber degrading strains (50 strains in total) belonged to Streptomyces group (33 strains, 66%) [45]. Streptomyces is a genus well known for its ability to secrete enzymes, which can metabolize or cleave biopolymers [3]. Interestingly, strains containing the second and third most lcp genes belonged to the mycolic acid producing genera (Mycobacteriodes and Nocardia).
The number of published family for Actinobacteria is 45 and 306 genera, leaving many more potential rubber degrading genera yet to be discovered . Among genera that could be of interest are thermophilic and mycolic acid strains such as Rhodococcus. In Table 2, 15 lcp genes from nine species were identified from Rhodococcus genus. To date, only two Rhodococcus strains able to degrade rubber have been isolated; R. rhodochrous strain RPK1 from a waste pond at a rubber processing factory in Thailand [45] and R. pyridinivorans strain F5 from samples collected from various contaminated rubber sites in Songkhla, Thailand [61]. Rhodococcus is a promising genus for rubber degradation as they are fast growers and easily cultivated. They have remarkably catabolic versatility, being able degrade an impressive array of xenobiotic and organic compounds. Presently, there are over 1800 and over 3200 patents retrieved for strains of this genus (Google patent search) respectively, as keywords [109].
Table 2. Diversity of Actinobacteria with DUF2236 domain containing lcp genes in GenBank database.
|
Family |
Genus |
Genus |
Species |
Lcp Genes |
Species |
|
Actinosynnemataceae |
1 |
Actinosynnema |
2 |
3 |
2 |
|
Corynebacterineae |
1 |
Williamsia |
3 |
6 |
2 |
|
Frankiaceae |
1 |
Frankia |
3 |
7 |
2 |
|
Gordoniaceae |
1 |
Gordonia |
12 |
23 |
2 |
|
Microbacteriaceae |
1 |
Microbacterium |
3 |
3 |
1 |
|
Micrococcaceae |
1 |
Psychromicrobium |
1 |
1 |
1 |
|
Micromonosporaceae |
2 |
Actinoplanes |
2 |
4 |
2 |
|
Micromonospora |
4 |
7 |
2 |
||
|
Mycobacteriaceae |
3 |
Mycobacterium |
3 |
6 |
2 |
|
Mycobacteroides |
5 |
43 |
9 |
||
|
Mycolicibacterium |
5 |
5 |
1 |
||
|
Nocardiaceae |
2 |
Nocardia |
12 |
29 |
2 |
|
Rhodococcus |
9 |
15 |
2 |
||
|
Nocardioidaceae |
2 |
Mumia |
1 |
3 |
3 |
|
Nocardioides |
4 |
4 |
1 |
||
|
Nocardiopsaceae |
2 |
Streptomonospora |
1 |
2 |
2 |
|
Nocardiopsis |
1 |
1 |
1 |
||
|
Pseudonocardiaceae |
7 |
Actinoalloteichus |
1 |
1 |
1 |
|
Amycolatopsis |
11 |
16 |
1 |
||
|
Kibdelosporangium |
1 |
2 |
2 |
||
|
Kutzneria |
1 |
1 |
1 |
||
|
Prauserella |
1 |
2 |
2 |
||
|
Pseudonocardia |
1 |
1 |
1 |
||
|
Saccharomonospora |
1 |
1 |
1 |
||
|
Streptomycetaceae |
1 |
Streptomyces |
39 |
51 |
1 |
|
Streptosporangiaceae |
2 |
Nonomuraea |
1 |
1 |
1 |
|
Streptosporangium |
1 |
1 |
1 |
||
|
Thermomonosporaceae |
1 |
Actinomadura |
2 |
2 |
1 |
|
Tsukamurellaceae |
1 |
Tsukamurella |
3 |
7 |
2 |
|
TOTAL |
29 |
|
134 |
248 |
|
Note: Species diversity = number of number of genes divided by number of species.
Thermotolerant Actinobacteria such as strains of Streptomyces albus and Streptomyces griseus, and thermophilic Actinobacteria such as Streptomyces, Amycolatopsis, Microbispora, Cellulosimicrobium, Micrococcus, Saccharopolyspora, Micromonospora, Thermobispora, Thermomonospora, Thermobifida, and Planomonospora were reported to be involved in the composting process [110]. A thermotolerant oil degrading Actinobacteria from the genera Rhodococcus and Gordonia showed ability to grow at up to 10% NaCl and utilize crude oil and individual hydrocarbons at higher (up to 50 °C) temperatures [111]. However, all rubber-degrading species described so far are mesophilic, with only one exception, identified as a Streptomyces sp. strain La 7, able to grow at 55 ˚C [112]. Eight moderate thermophilic strains (50 °C) were isolated and tentatively identified as Actinomadura sp.*, Nocardia farcinica, and Thermomonospora curvata (blast homology 96.5*–99.9%) [86].
Among the deposited Actinobacteria strains containing lcp genes, 86 of them were isolated from various sources across the world, mainly from soil samples (57%), plant samples (20%), clinical samples (14%), water samples (7%), and animal/ insect samples (2%). Clinical samples mainly consist of Nocardia, Mycobacterium, and Mycobacteroides strains (Supplementary Table S1). Lcp containing strains were also isolated from plants that do not produce latex, such as wheat plants and oak trees.
The conserved region for the strains isolated from various sources showed high similarity to the region reported by Rother et al. (2016) (KTRLVHAAVRHLL) (see Section 4.1) except for a sample from animal origin (RVRLIHGLVRKHV) [76]. The strain Mycolicibacterium malmesburyense (CRL78180) was isolated from cattle (Bos Taurus) in South Africa. When comparing the 248 sequences from these samples, there are other highly conserved regions to be further examined (Supplementary Figure S2). Further investigation of these conserved regions would likely identify novel functions attributed to these strains.
7. Rubber Biodegradation
Researchers have been studying microbial rubber degradation since the early 1900s. In this section, we discuss the main methods used to conduct and evaluate rubber degradation under laboratory conditions and possible ways to apply rubber biodegradation in our current regime.
7.1. In vivo Rubber Degradation Using Microbial Culture
In the laboratory, rubber degradation studies are carried out by incubating the microorganism with rubber materials (0.5–0.6% w/v) supplemented with mineral salts medium (MSM) on a shaker for 6–12 weeks at 30 °C [64][113]. Examination of the size, final mass, and morphology of the rubber materials after incubation indicates the relative activity of the microorganism in question. Alternatively, the bacteria are inoculated on NR latex (concentrated and purified) overlay agar plates, consisting of a bottom agar layer of MSM and incubated for 3–7 days at 30 °C. Their ability to form clear zones on the NR latex layer are then evaluated.
Table 3 shows the rate of rubber degradation for some strains. Among the parameters recorded were duration, percent weight loss, material tested and pre-treatment before introducing the bacteria. Most of the studies were carried out at 150 rpm shaking. Separate observations showed the unfavorable effects of agitation on the rate of rubber degradation. For example, the formation of biofilms could be limited due to constant agitation. Nocardia sp. 835A-Rc, a mutant strain, uniformly attacked tyre particles when stirred at 0–40 rpm. However, higher stirring rates led to some clumps of microbial colonies on the rubber surface and separate deep semi-spherical cavities [114]. Using G. polyisoprenivorans VH2 culture, Berekaa and his colleagues (2000) tested six weeks of shaking, followed by six weeks of stationary incubation, which led to the complete disintegration of the rubber material [113].
Table 3. Compilation of in vivo culture testing on NR degrading.
|
Culture |
Duration (Weeks) |
Weight Loss (%) |
Material |
Pre-Treatment |
Reference |
|
Nocardia sp. strain 835A |
8 |
100 |
NR |
– |
[40] |
|
90 |
Latex glove |
Acetone & CHCl3 |
|||
|
S. cummioxidans 35Y |
1 |
60 |
NR |
NS |
[46] |
|
Amycolatopsis S1A |
6 |
11 |
Rubber coated slides (absence of organic nutrients) |
Soxhlet purification |
[115] |
|
Amycolatopsis S1D |
12 |
||||
|
Nocardia S3F |
13 |
||||
|
Streptomyces S1G |
44 |
||||
|
Streptomyces S3D |
21 |
||||
|
Streptomyces S4C |
26 |
||||
|
Streptomyces S4D |
43 |
||||
|
Streptomyces S4E |
37 |
||||
|
Streptomyces S4F |
43 |
||||
|
Streptomyces S4G |
38 |
||||
|
S. sp. strain LA7 |
8.6 |
80 |
Emulgated latex |
Dialysed latex |
[112] |
|
3.3 |
60 |
||||
|
10 |
29.4 |
Latex glove |
Acetone & CHCl3 |
||
|
10 |
31.3 |
Latex glove + Triton X (0.1%, w/w) |
Acetone & CHCl3 |
||
|
S. coelicolor CH13 |
6 |
14 |
Latex glove |
– |
[116] |
|
92 |
Latex glove + starch (65%) |
NS |
|||
|
Nocardia sp. strain 385A |
10 |
5.5 |
Fresh latex |
NS |
[117] |
|
Bacillus cereus |
2 |
3.5 |
Tyre |
NS |
[118] |
|
7.8 |
NR |
Acetone |
|||
|
12.3 |
Latex film of MR + metroxylan sago pith waste |
||||
|
R. pyridinivorans strain F5 |
4 |
9.36 |
Latex glove |
– |
[60] |
NS: not stated, – : none
Pre-treatment of samples to remove impurities, composites materials and antimicrobial substances improved colonization efficiency and degradation rates [113]. The impurities removed by pre-treatment can otherwise leach into the surrounding media during long cultivation periods and may be unfavorable to microbial growth. Constant replacement of MSM media also enhanced the rates rubber disintegration which then led to complete visual loss of the rubber material at week 10 [113].
The addition of a starch component to the incubation media also improved degradation. The starch is metabolized first and acts as a growth substrate for rubber degrading bacteria, to increase initial bacterial populations rapidly, and subsequently the rate of NR degradation [116][117][118][119][120].
Actinobacteria has also showed their ability to degrade IR breaking cross-link Sulfur-Sulfur or Carbon-Sulfur bonds with the main chain remaining intact. Streptomyces sp. was able to devulcanize IR after 4 weeks incubation, having 18.13% of carbon loss and 52.90 reduction in cross linking degrees [121]. Streptomyces coelicolor 1A was able to degrade IR (Aldrich, Mw 800,000), having 9% of IR detected with Mw less than 10,000 after six weeks [64]. Gordonia amicalisa was able to reduce vulcanized IR and vulcanized styrene butadiene rubber crosslink densities by 13.7% and 22.1%, respectively, vulcanized styrene butadiene rubber sulfur content on its surface was decreased by 22.9%, S-S bonds were broken, producing S=O bonds [122]. Amycolaptopsis sulphurea DSMZ 46092, Gordonia DSMZ 44215 and Gordonia DSMZ 44369 showed 20.90%, 18.64%, and 17.52% respectively showed decrease in sulfur after 4 weeks of incubation [123]. In patent US 7737191B2, mycolic acid containing Actinobacteria strains of Corynebacterium, Rhodococcus, Nocardia, Gordonia, Tsukamurella, Dietzia, and Mycobacterium, and mainly Gordonia desulfuricans strain SG213E were used for rubber treatment [124].
These findings show the potential of Actinobacteria to degrade both NR and IR in treatment of rubber wastes.
7.1.1. Measurements for Rubber Degradation
The efficiency of bacteria in degrading rubber is evaluated based on the organism’s effect on several stages of biodegradation: biodeterioration, biofragmentation, bioassimilation and mineralization. Scanning electron microscopy (SEM) is mainly used to observe the degree of deterioration that the rubber material undergoes following exposure to the organism. In the main, this is a qualitative measure that looks for changes in the surface of the rubber.
In biofragmentation, the original polymer is fragmented resulting in fewer cis-1,4 double bonds, and the formation of new carbonyl groups. These chemical changes are usually detected using attenuated Total Reflection-Fourier transform infrared (ATR-FTIR) spectra. Based on the ATR-FTIR profile for NR, changes in ~873 cm−1 indicate change of C=H bond, double peak at 1250–1500 cm−1 refers to the presence of amide I and II, while at ~1715 cm−1 peak refer to C=O bond, and ~3400 cm−1 peak refer to O=H bond [125]. Presence of aldehydes containing oligomers can also be tested by staining the rubber material using Schiff’s reagent, resulting in purple coloration.
Differences in rubber molecular weight before and after bacterial inoculation, can be measured using Gel Permeation Chromatography (GPC). Weight loss measurements indicate that the bacteria are able to cleave the carbon backbone of NR or IR and utilize the low molecular-weight degradation products for growth. Polymer without microbial inoculation (control) shows Gauss-like distribution of molecular weights between 104 and 107 (Mw, 8 × 105) [64].
The rate of rubber mineralization under aerobic condition can be calculated by measuring the amount of CO2 released during the cultivation of cells with rubber materials in tightly closed Erlenmeyer flasks over time. The released CO2 is trapped as BaCO3, from reaction with Ba(OH)2, which is subsequently processed to determine the CO2 content [116].
For IR, additional measurements can be carried out. Scanning Electron Microscopy (SEM) coupled to Energy Dispersive Spectroscopy (EDS) will enable comparison of EDS maps for carbon (C), oxygen (O) and sulfur (S) with control samples [121]. Horikx analysis determines the breakdown in a vulcanized rubber network by the rate of increase of the soluble fraction of the rubber as a function of the measured cross-link density of the remaining total insoluble fraction is different for cleavage of S-S and C-S bonds [126]. Crosslinking degree measurements can be determined by boiling 20 mg of vulcanized rubber in a tightly closed filter paper at 110.6 °C to a leaching system for 48 h, the crosslinking degree, related to the insoluble residue, was calculated as the ratio between the weights of the sample after and before being soaked in toluene during the leaching process [121].
7.1.2. Drawbacks
The quantitative measurement of in vivo rubber degradation using culture is limited and the results from studies on different strains are often difficult to compare. The presence of clearing zones, rates of growth, and zone sizes on NR latex overlay agar plates are affected by various parameters such as strain type, growth rate, strain age, amount of strain inoculated (a drop, a colony etc.). Several of the most potent rubber degraders (G. polyisoprenivorans and G. westfalica) do not produce clearing zones and requires close contact with the rubber material they degrade. Methods employed for rubber degradation studies are also inconsistent between different research groups (type, size and thickness of rubber material, rubber material pre-treatment, number of days, amount of inoculum, agitation rate etc.). The surface characteristics of the tested material are crucial as high surface ratio (smaller particles) and the presence of rough rubber surfaces and cracks are favourable for microorganism growth and attachment [138]. SEM observation showed that Paecilomyces variotii strain SFA-25 hyphal network and rubber surface erosion was more intense on rough and cracked surfaces compared to smooth surfaces.
Rubber degrading strains may contain more than 1 lcp gene, combination of the gene products may have synergistic activity; alternatively, the products may be dysfunctional. Further investigations into the functions of additional genes, especially in the chromosome, are needed to understand how these lcp genes may or may not interact to degrade rubber. Three types of rubber oxygenases for the enzymatic cleavage of rubber has been tested using RoxAXsp, RoxBXsp, LcpK30 or with combinations of the three proteins resulting in differed in the number of intact isoprene units, having the same general structure with terminal functions (CHO-CH2- and -CH2-COCH3) [48].
7.2. In situ Rubber Degradation
Several possible strategies for using Actinobacteria for in situ rubber degradation are discussed here: bioaugmentation, biotransformation, bioreactors, and synthetic biology. There is a strong possibility that combining these strategies with the appropriate strain could be applied for use at larger scale to help reduce rubber pollution in the environment.
7.2.1. Bioaugmentation and Consortia
Biomimicry can improve the degradation process through introduction of efficient microbial strains (bio-augmentation) or by addition of nutrients to the soil (bio-stimulation) [127]. Bioaugmentation is a technique for improving the degradative capacity of contaminated areas through introduction of specific competent strains or consortia of microorganisms (autochthonous or allochthonous wild type or genetically modified microorganisms, GEMs) [128].
The main limitation of using bioaugmentation for rubber wastewater treatment is the microbial survival rate. Several potential solutions can be explored such as increasing the amount of inoculant or incorporation of inoculant by time-based, addition of nanomaterials to support the organism, and the of GEMs to improve survival [129].
Bio-stimulation refers to the addition of growth-rate limiting nutrients like phosphorus, nitrogen, oxygen, electron donors to severely polluted sites for the enhancing the indigenous biomass activity to degrade the hazardous and toxic contaminants [127]. The primary advantage of bio-stimulation is that bioremediation will be undertaken by already present native microorganisms that are well-suited to the subsurface environment and are well distributed spatially within the subsurface [130].
Recently, the use of monoculture, co-culture and consortium in rubber degradation studies, mimicking the natural environmental condition, has been compared [50][60][68][130][131]. The degradation of complex molecules is achieved through co-operative degradation where one organism transforms the original material into products that can be utilized by the other organisms [50][132]. The synergetic interaction of several strains of microorganisms in rubber degradation activity was better when compared to individual strains. This strategy where bacterial strains are combined to enhance the overall degradation of rubber is worth further investigation to understand what combinations work best.
7.2.2. Biotransformation
Microbial fermentation has the potential for making three renewable rubber intermediates: isoprene, isobutene, and butadiene [15]. However, these processes must be cost competitive with the petrochemical pathways. The enzyme isoprene synthase has been identified in plants and through synthetic biology its expression has been optimized in several microorganisms. Andler et al. (2019) proposed a recycling method for rubber waste through biotransformation [133]. Carbon sources obtained from the biodegradation of rubber were used to produce Polyhydroxyalkanoates (PHAs) through recombinant strains of G. polyisoprenivorans VH2 harboring plasmid pAK68 (phaCAB from Ralstonia eutropha) and pAK71 (phaC1 from Pseudomonas aeruginosa). PHAs are environmentally friendly, biodegradable alternatives to petroleum-based plastics made by a group of bacterial polyesters.
There has been success in improving rubber degradation by using a mutant strain Nocardia 835A strain Rc, which caused 81% weight loss to tyre thread after eight weeks and its activity was influenced by the NR content [65].
7.2.3. Bioreactors
The use of bacterial strains in rubber wastewater treatment is considered an ecologically and environmentally favorable technology, it provides many potential advantages, such as low energy consumption, ease of the process, low equipment requirements, and no pollution [134]. Stevenson et al. (2008) and other scientists recommend a multistage process utilizing several different microbes and biochemical pathways [135]. Here, the detoxification of scrap tyre rubber (e.g., fungus Recinicium bicolour) is followed by sulfur-oxidization (e.g., bacteria Thiobacillus ferroxidans or sulfur-reducing archaeon (Pyrococcus furiosus). They propose the usage of Actinobacteria for rubber mineralisation and catabolism based on the production of lcp and rox enzymes after the waste materials have been detoxified and devulcanized. Actinobacteria strains such as Rhodococcus rhodochrous, Arthrobacter sulfureus, Gordonia rubropertincta, and Rhodococcus erythropolis can also play a role in desulfurizing [136]. The most common biodesulfurization pathway reported to date is the 4S pathway discovered in Rhodococcus erythropolis IGTS8 [137].
7.2.4. Synthetic Biology
Actinobacteria demonstrate many advantages as a rubber degrader, however they can be further improved for application purposes, whether in the industrial process or the environment through synthetic biology. Synthetic biology uses a pared-down life form to serve as a chassis on which to build something with a useful application to humankind such as biodegradation, recycling, waste clean-up, etc. [138]. Only ten microbes have been “domesticated” for industrial use [139]. Synthetic biologists develop their projects through standard engineering cycles of ‘design, build, test’ [138]. The design phase involves computer modelling of the components’ behavior. The build stage involves the genetic engineering. The test step assesses whether the prospective approach works.
Cell-free protein synthesis is another powerful flexible bottom-up approach utilizing a minimum of cellular elements that allows for labor- and time-efficient protein expression in a test tube without multistep complex maintenance of a living culture. The move to cell-free synthesis can increase reaction rates 100-fold [140]. Karzbrun et al. (2014) assembled two-dimensional DNA compartments fabricated in silicon capable of metabolism, programmable protein synthesis, and communication as a powerful route for flexible and controllable production systems [141]. This approach can be used to access non-natural materials and circumvents the potential issue of substrate toxicity to cells (as well as the regulatory issues of genetically modified cells [139].
Building blocks for IR (such as isoprene) are currently sourced from petroleum. Synthetic biology would reduce the cost, carbon footprint of tyres, and most importantly, addresses the issues of microplastics pollution in the environment. Scientists in China have altered a marine bacterium Synechococcus elongatus photosynthesis pathway to produce high quantities of isoprene in the laboratory, while researchers in the US are using yeast to convert plant carbohydrates into low-cost alcohols and later converted into isoprene [142][143].
7.2.5. Drawbacks
Rubber biodegradation has yet to be applied outside laboratory testing or at a large scale. Small scale studies involving soil, surface water and groundwater, showed that introduced microorganisms are unable to survive in rubber liquid wastes. There may be other unknown limitations in biodegradation using living cultures or enzymes that we have yet to discover.
There is, however, one successful full scale bioaugmentation story that has been reported is the in in-situ removal of chlorinated solvents in ground water, with the use of anaerobic bacteria of Dehalococcoides group [144].
In rubber biodegradation, Actinobacteria has many of advantages however one must be aware that there are strains which pathogenic towards animals (R. equi), plants (R. fascians), and humans (N. farcinia, R. equi, R. rhodochrous, and R. erythropolis). In situ application of living strains and GEMs would require environmental impact assessment and further evaluation.
7.3. Rubber Reclamation of Tyres by Microbes
Currently, there are vast tyre landfill areas to be addressed, and we can start improving the rate of tyre degradation by introducing beneficial microorganisms including rubber degrading strains to these areas. One must start by enriching and acclimatizing the culture to the landfill or providing suitable starter material, like techniques used in composting. The existing tyres must be reduced in size and pretreated in order to facilitate microbial degradation.
In addressing rubber waste problems, we must also find ways to make rubber products such as tyres using materials that are easier to degrade. Researchers found that using ruthenium to generate cyclopentene as a synthetic rubber material are completely degradable at lower temperature (40–50 °C) [145]. Michelin has proposed the usage of 3-D printing using biodegradable materials for their tyre production [146]. Goodyear recently unveiled a tyre concept ‘reCharge’ using dandelion rubber and fibers that can generate tyre thread as needed [147]. Biodegradation of these materials can then be more feasible in an industrial context (recycling facility, bioreactor) or at landfills.
8. Concluding Remarks
In this present review, we have highlighted the incredible utility of rubber in the production of a diverse range of commercial products. Actinobacteria is a group of microorganisms that have the potential to be developed as part of an industrial solution for the biodegradation of NR and IR. Actinobacteria are present in a diverse range of environments, indicating their ability to adapt to local surroundings and thus a survival advantage in withstanding industrial conditions. Of more importance, the strains of Actinobacteria that express dioxygenases which are implicated in the degradation of rubber. Although the enzyme activities were discovered decades ago, it is only with the convergence of genomic screening with functional tests that the distribution and evolutionary patterns of the genes responsible for the enzymes are being elucidated. Molecular techniques are now being used to confirm the mechanisms of action behind rubber degradation by these organisms. Our present understanding of how the dioxygenases act on rubber substrates, and what bacterial, molecular, or environmental factors best support their activity is limited, and it will be crucial to continue screening for novel strains or gene products in tandem with functional assays to fully understand the mechanistic steps behind rubber degradation by the Actinobacteria, and the range of activity they are capable of.
Identifying the critical steps and environmental or molecular factors in the degradation of rubber by Actinobacteria will allow the rational design of bio-processes that can be used in industrial settings. Synthetic biology will be important for producing enzyme stocks, identifying enzyme combinations that degrade rubber synergistically, or integrating active components onto carriers that are designed to protect the active components from harsh industrial conditions. In conclusion, identifying strains of Actinobacteria that degrade rubber is the first step in developing processes for bio-degrading rubber tyres. The second step is to understand the steps and components that play pivotal roles in the process, and to use this knowledge to design bio-systems that can be scaled up and used under industrial conditions.
Supplementary Materials: The following are available online at https://www.mdpi.com/article/10.3390/polym13121989/s1, Figure S1: Phylogenetic clusters (12) of 248 lcp genes obtained from GenBank, Figure S2: Conserved regions of 248 lcp amino acid sequences from GenBank. Sequences were viewed using Jalview, dark purple shades indicate highest percentage identity among sequences, Table S1: Details of Isolation Source for Actinobacteria containing Lcp Gene in GenBank.
Author Contributions: Conceptualization, A.A.B, J.-J. S. and K.S.; writing—original draft preparation, A.A.B; writing—review and editing, J.-J.S. and K.S; supervision, K.S. and C.T.Y; project administration, K.S. and C.T.Y; funding acquisition, A.A.B. and C.T.Y. All authors have read and agreed to the published version of the manuscript.
Funding: The main author is funded by the Government of Sarawak through a PhD scholarship under USM grant number 304.PBIOLOGI.6501009.J136.
Institutional Review Board Statement: Not applicable.
Informed Consent Statement: Not applicable.
Acknowledgments: We would like to thank Sarawak Biodiversity Centre (SBC) and Ecobiomaterial Research Laboratory (USM) members for their support.
Conflicts of Interest: The authors declare no conflict of interest.
[80][148][1][54][149][150][2][3][4][5][6][7][8][9][10][11][12][13][14][15][16][17][18][19][20][21][22][23][24][25][26][27][28][29][30][31][32][33][34][35][36][37][38][39][40][41][42][43][44][45][46][47][48][49][50][51][52][53][55][56][57][58][59][60][61][62][63][64][65][66][67][68][69][70][71][72][73][74][75][76][77][78][79][81][82][83][84][85][86][87][88][89][151][152][90][91][153][98][92][154][155][93][94][95][96][97][99][100][101][102][103][104][156][105][106][107][108][157][158][109][110][111][112][113][114][115][116][117][118][119][120][121][122][123][124][125][126][159][127][128][129][130][131][132][133][134][135][136][137][138][139][140][141][142][143][144][145][146][147]
References
- Birke, J.; Jendrossek, D. Rubber Oxygenase and Latex Clearing Protein Cleave Rubber to Different Products and Use Different Cleavage Mechanisms. Appl. Environ. Microbiol. 2014, 80, 5012–5020.
- Tanaka, Y.; Sakdapipanich, J.T. Chemical Structure and Occurrence of Natural Polyisoprenes. Biol. Chem. Biotechnol. Appl. 2005, 2.
- Shah, A.A.; Hasan, F.; Shah, Z.; Kanwal, N.; Zeb, S. Biodegradation of natural and synthetic rubbers: A review. Int. Biodeterior. Biodegrad. 2013, 83, 145–157.
- Ilcu, L.; Röther, W.; Birke, J.; Brausemann, A.; Einsle, O.; Jendrossek, D. Structural and Functional Analysis of Latex Clearing Protein (Lcp) Provides Insight into the Enzymatic Cleavage of Rubber. Sci. Rep. 2017, 7, 1–11.
- Backhaus, R.A. Rubber Formation in Plants—A Mini-Review. Isr. J. Bot. 1985, 34, 283–293.
- Jendrossek, D.; Birke, J. Mini-Review Rubber Oxygenases. Appl. Microbiol. Biotechnol. 2018, 103, 125–142.
- Rose, K.; Tenberge, K.B.; Steinbüchel, A. Identification and Characterization of Genes from Streptomyces sp. Strain K30 Responsible for Clear Zone Formation on Natural Rubber Latex and Poly(cis-1,4-isoprene) Rubber Degradation. Biomacromolecules 2005, 6, 180–188.
- Yikmis, M.; Steinbüchel, A. Historical and Recent Achievements in the Field of Microbial Degradation of Natural and Synthetic Rubber. Appl. Environ. Microbiol. 2012, 78, 4543–4551.
- Cornish, K. Alternative Natural Rubber Crops: Why Should We Care? Technol. Innov. 2017, 18, 244–255.
- Brüning, K. Natural Rubber. In Chemistry and Materials Science; Kobayashi, S.M.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2014.
- Puskas, J.E.; Chiang, K.; Barkakaty, B. Natural Rubber (NR) Biosynthesis: Perspectives from Polymer Chemistry. In Chemistry, Manufacture and Applications of Natural Rubber; Elsevier Inc.: Amsterdam, The Netherlands, 2014; pp. 30–67.
- Warneke, S.; Arenskötter, M.; Tenberge, K.B.; Steinbüchel, A. Bacterial degradation of poly(trans-1,4-isoprene) (gutta percha). Microbiology 2007, 153, 347–356.
- OECD. Emission Scenario Document on Additives in Rubber Industry. Available online: (accessed on 4 June 2020).
- White, J.L. First of a Series: Pioneering Polymer Industry Developments: Bayer and the First Synthetic Rubber First of a Series. Int. Polym. Process. 1999, 14, 114.
- Statisca. Consumption of Natural and Synthetic Rubber Worldwide from 1990–2018 (in 1000 Metric Tons). Available online: (accessed on 12 March 2021).
- The Synthetic Rubber Production Process. AQUASEAL Rubber Limited. Available online: (accessed on 4 June 2020).
- Tiseo, I. Rubber—Statistics; Facts. Available online: (accessed on 13 March 2021).
- Andrew, L. How Long it Takes 50 Common Items to Decompose. Available online: (accessed on 24 July 2020).
- Clark, T. Enhancing the Biodegradation of Waste Rubber Discarded Rubber Materials. International Latex Conference 2015. Available online: (accessed on 24 July 2020).
- Unciano, N.M. Microbial Processing of Natural Rubber Waste. Insight in the Microbial Technologies and Methods for Bioremediation. Available online: (accessed on 19 March 2020).
- Ward, J. Synthetic Biology: From Science to Bioeconomy Application: Opportunities for Challenge-led Innovation. Available online: (accessed on 13 March 2020).
- Williams, P.T. Pyrolysis of Waste Tyres: A Review. Waste Manag. 2013, 33, 1714–1728.
- EPA. Rubber and Leather: Material-Specific Data. Agency, United States Environmental Protection. Available online: (accessed on 30 April 2020).
- WBSCD. Development for WBSCD. Stakeholders Launch Global Platform for Sustainable Natural Rubber. Available online: (accessed on 2 April 2019).
- Oriaku, E.C.; Agulanna, C.N.; Odenigbo, J.; Nnoruka, N. Waste to Wealth through the Incineration of Waste Tyres and Recovery of Carbon Black. Int. J. Multidiscip. Sci. Eng. 2013, 4, 30–36.
- Makitan, V. Waste Tyre Recycling: Current Status, Economic Analysis and Process Development. Ph.D. Thesis, Curtin University, Bentley, Australia, 2010. Available online: (accessed on 13 March 2021).
- EPA. Air Emissions from Scrap Tire Combustion. Agency, United States Environmental Protection. Available online: (accessed on 13 March 2019).
- Wądrzyk, M.; Janus, R.; Rządzik, B.; Lewandowski, M.; Budzyń, S. Pyrolysis Oil from Scrap Tires as a Source of Fuel Components: Manufacturing, Fractionation, and Characterization. Energy Fuels 2020, 34, 5917–5928.
- Yong, Z.J.; Bashir, M.J.; Ng, C.A.; Sethupathi, S.; Lim, J.W.; Show, P.L. Show Sustainable Waste-to-Energy Development in Malaysia: Appraisal of Environmental, Financial, and Public Issues Related with Energy Recovery from Municipal Solid Waste. Processes 2019, 7, 676.
- EPA. Tire Fires. Agency, United States Environmental Protection. Available online: (accessed on 13 March 2019).
- A Study on Scrap Tyres Management for Peninsular Malaysia. Final Report (September 2011). Chemsain Konsultant Sdn. Bhd. Available online: (accessed on 13 March 2020).
- Holdings, D. Waste Tyre Solution. Available online: (accessed on 30 April 2020).
- Halle, L.L.; Palmqvist, A.; Kampmann, K.; Khan, F.R. Ecotoxicology of micronized tire rubber: Past, present and future considerations. Sci. Total. Environ. 2020, 706, 135694.
- Chatterjee, S.; Sharma, S. Microplastics in Our Oceans and Marine Health. Field Actions Sci. Reports. J. Field Actions 2019, 19, 54–61.
- Tamma, P. Tires Tread on the Environment. Available online: (accessed on 12 March 2020).
- Sieber, R.; Kawecki, D.; Nowack, B. Dynamic probabilistic material flow analysis of rubber release from tires into the environment. Environ. Pollut. 2020, 258, 113573.
- TheStar. Car Tyres a Likely Source of Microplastics in Coastal Waters. Available online: (accessed on 12 March 2020).
- Wiess, K.R. The Pileup of Plastic Debris Is More than Ugly Ocean Litter. Available online: (accessed on 12 March 2020).
- UEG. UEG Week: Microplastics Discovered in Human Stools across the Globe in First Study of Its Kind. Gastroenterology, United European. Available online: (accessed on 30 April 2020).
- Serumgard, J. Internalization of Scrap Tire Management Costs: A Review of the North American Experience. In Rubber and the Environment, Proceedings of the Joint Workshop of the Secretariat of the United Nations Conference on Trade and Development and the International Rubber Study Group on Rubber and the Environment, Bali, Indonesia, 30 October 1998; United Nations Conference on Trade and Development: Geneva, Switzerland.
- Montazer, Z.; Najafi, M.B.H.; Levin, D.B. Challenges with Verifying Microbial Degradation of Polyethylene. Polymers 2020, 12, 123.
- Joutey, N.T.; Bahafid, W. Biodegradation: Involved Microorganisms and Genetically Engineered Microorganisms. Biodegrad. Life Sci. 2013, 289–320.
- Taysum, D.H. In Microbiological Deterioration in the Tropics. In Monograh No. 23; Society of Chemical Industry: London, UK, 1966.
- Kasai, D. Poly(cis-1,4-isoprene)-cleavage enzymes from natural rubber-utilizing bacteria. Biosci. Biotechnol. Biochem. 2020, 84, 1089–1097.
- Tsuchii, A.; Suzuki, T.; Takeda, K. Microbial Degradation of Natural Rubber Vulcanizates. Appl. Environ. Microbiol. 1985, 50, 965–970.
- Luo, Q.; Hiessl, S.; Poehlein, A.; Daniel, R.; Steinbüchel, A. Insights into the Microbial Degradation of Rubber and Gutta-Percha by Analysis of the Complete Genome of Nocardia nova SH22a. Appl. Environ. Microbiol. 2014, 80, 3895–3907.
- Gibu, N.; Arata, T.; Kuboki, S.; Linh, D.V.; Fukuda, M.; Steinbüchel, A.; Kasai, D. Characterization of the genes responsible for rubber degradation in Actinoplanes sp. strain OR16. Appl. Microbiol. Biotechnol. 2020, 104, 7367–7376.
- Röther, W.; Birke, J.; Grond, S.; Beltran, J.M.; Jendrossek, D. Production of functionalized oligo-isoprenoids by enzymatic cleavage of rubber. Microb. Biotechnol. 2017, 10, 1426–1433.
- Watcharakul, S.; Röther, W.; Birke, J.; Umsakul, K.; Hodgson, B.; Jendrossek, D. Biochemical and spectroscopic characterization of purified Latex Clearing Protein (Lcp) from newly isolated rubber degrading Rhodococcus rhodochrous strain RPK1 reveals novel properties of Lcp. BMC Microbiol. 2016, 16, 1–13.
- Jendrossek, D.; Tomasi, G.; Kroppenstedt, R.M. Bacterial Degradation of Natural Rubber: A Privilege of Actinomycetes? FEMS Microbiol. Lett. 1997, 150, 179–188.
- Tsuchii, A.; Takeda, K. Rubber-Degrading Enzyme from a Bacterial Culture. Appl. Environ. Microbiol. 1990, 56, 269–274.
- Linos, A.; Berekaa, M.M.; Reichelt, R.; Keller, U.; Schmitt, J.; Flemming, H.-C.; Kroppenstedt, R.M.; Steinbüchel, A. Biodegradation of cis-1,4-Polyisoprene Rubbers by Distinct Actinomycetes: Microbial Strategies and Detailed Surface Analysis. Appl. Environ. Microbiol. 2000, 66, 1639–1645.
- Arenskötter, M.; Bröker, D.; Steinbüchel, A. Biology of the Metabolically Diverse Genus Gordonia. Appl. Environ. Microbiol. 2004, 70, 3195–3204.
- Hiessl, S.; Böse, D.; Oetermann, S.; Eggers, J.; Pietruszka, J.; Steinbüchel, A. Latex Clearing Protein—An Oxygenase Cleaving Poly(cis-1,4-Isoprene) Rubber at the cis Double Bonds. Appl. Environ. Microbiol. 2014, 80, 5231–5240.
- Watcharakul, S. Isolation of a Novel Rubber Degrading Bacterium from a Consortium and Characterization of Its lcp Gene Products Sirimaporn Watcharakul A Thesis Submitted in Fulfillment of the Requirements for the Doctor of Philosophy in Microbiology. Ph.D. Thesis, Prince of Songkla University, Songkhla, Thailand, 2017.
- Altenhoff, A.-L.; de Witt, J.; Andler, R.; Steinbüchel, A. Impact of additives of commercial rubber compounds on the microbial and enzymatic degradation of poly(cis-1,4-isoprene). Biodegradation 2019, 30, 13–26.
- Yikmis, M.; Arenskӧtter, M.; Rose, K.; Lange, N.; Wernsmann, H.; Wiefel, L.; Steinbüchel, A. Secretion and Transcriptional Regulation of the Latex-Clearing Protein, Lcp, by the Rubber-Degrading Bacterium Streptomyces sp. Strain K30. Appl. Environ. Microbiol. 2008, 74, 5373–5382.
- Yikmis, M.; Steinbüchel, A. Importance of the Latex-clearing protein (Lcp) for Poly(cis-1,4-isoprene) Rubber Cleavage in Streptomyces sp. K30. Microbiol. Open 2012, 1, 13–24.
- Brӧker, D.; Arenskӧtter, M.; Legatzki, A.; Nies, D.H.; Steinbüchel, A. Characterization of the 101-Kilobase-Pair Megaplasmid pKB1, Isolated from the Rubber-Degrading Bacterium Gordonia westfalica Kb1. J. Bacteriol. 2004, 186, 212–225.
- Hiessl, S.; Schuldes, J.; Thürmer, A.; Halbsguth, T.; Bröker, D.; Angelov, A.; Liebl, W.; Daniel, R.; Steinbüchel, A. Involvement of Two Latex-Clearing Proteins during Rubber Degradation and Insights into the Subsequent Degradation Pathway Revealed by the Genome Sequence of Gordonia polyisoprenivorans Strain VH2. Appl. Environ. Microbiol. 2012, 78, 2874–2887.
- Linh, D.V.; Huong, N.L.; Tabata, M.; Imai, S.; Iijima, S.; Kasai, D.; Anh, T.K.; Fukuda, M. Characterization and functional expression of a rubber degradation gene of a Nocardia degrader from a rubber-processing factory. J. Biosci. Bioeng. 2017, 123, 412–418.
- Brӧker, D.; Dietz, D.; Arenskӧtter, M.; Steinbüchel, A. The Genomes of the Non-Clearing-Zone-Forming and Natural-Rubber- Degrading Species Gordonia polyisoprenivorans and Gordonia westfalica Harbor Genes Expressing Lcp Activity in Streptomyces Strains. Appl. Environ. Microbiol. 2008, 74, 2288–2297.
- Birke, J.; Jendrossek, D. Solimonas fluminis has an active latex-clearing protein. Appl. Microbiol. Biotechnol. 2019, 103, 8229–8239.
- Nawong, C.; Umsakul, K.; Sermwittayawong, N. Rubber gloves biodegradation by a consortium, mixed culture and pure culture isolated from soil samples. Braz. J. Microbiol. 2018, 49, 481–488.
- Jendrossek, D.; Reinhardt, S. Sequence Analysis of a Gene Product Synthesized by Xanthomonas sp. during Growth on Natural Rubber Latex. FEMS Microbiol. Lett. 2003, 224, 61–65.
- Sharma, V.; Siedenburg, G.; Birke, J.; Mobeen, F.; Jendrossek, D.; Prakash, T. Metabolic and Taxonomic Insights into the Gram-negative Natural Rubber Degrading Bacterium Steroidobacter cummioxidans sp. nov., strain 35Y. PLoS ONE 2018, 13, 1–20.
- Birke, J.; Röther, W.; Jendrossek, D. Rhizobacter gummiphilus NS21 has two rubber oxygenases (RoxA and RoxB) acting synergistically in rubber utilisation. Appl. Microbiol. Biotechnol. 2018, 102, 10245–10257.
- Bode, H.B.; Zeeck, A.; Pluckhahn, K.; Jendrossek, D. Physiological and Chemical Investigations into Microbial Degradation of Synthetic Poly(cis-1,4-isoprene). Appl. Environ. Microbiol. 2000, 66, 3722–3726.
- Chengalroyen, M.D.; Dabbs, E.R. The Biodegradation of Latex Rubber: A Mini review. J. Polym. Environ. 2013, 21, 874–880.
- Imai, S.; Ichikawa, K.; Kasai, D.; Masai, E.; Fukuda, M. Isolation and characterization of rubber-degrading bacteria. J. Biotechnol. 2010, 150, 237.
- Kanwal, N.; Shah, A.A.; Qayyum, S.; Hasan, F. Optimization of pH and temperature for degradation of tyre rubber by Bacillus sp. strain S10 isolated from sewage sludge. Int. Biodeterior. Biodegrad. 2015, 103, 154–160.
- Krishnaswamy, V.; Ahongsangbam, N.A. Study on Mineralisation of Poly(cis-1,4-isoprene) and Synthetic Rubber Gloves (SRG) by the Bacterial Consortium. Ann. Appl. Microbiol. Biotechnol. J. 2017, 1, 1–10.
- Muralidharan, M.; Krishnaswamy, V. Artificial rubber mineralization by co-cultured bacterial strains. Int. J. Biol. Res. 2016, 4, 105.
- Birke, J.; Röther, W.; Schmitt, G.; Jendrossek, D. Functional Identification of Rubber Oxygenase (RoxA) in Soil and Marine Myxobacteria. Appl. Environ. Microbiol. 2013, 79, 6391–6399.
- Enoki, M.; Doi, Y.; Iwata, T. Oxidative Degradation ofcis-andtrans-1,4-Polyisoprenes and Vulcanized Natural Rubber with Enzyme-Mediator Systems. Biomacromolecules 2003, 4, 314–320.
- Rose, K.; Steinbüchel, A. Biodegradation of Natural Rubber and Related Compounds: Recent Insights into a Hardly Understood Catabolic Capability of Microorganisms. Appl. Environ. Microbiol. 2005, 71, 2803–2812.
- Fetzner, S. Oxygenases without requirement for cofactors or metal ions. Appl. Microbiol. Biotechnol. 2002, 60, 243–257.
- Mohamed, N.H.; Ismail, M.A.; Abdel-Mageed, W.M.; Shoreit, A.A. Biodegradation of Natural Rubber Latex of Calotropis procera by Two Endophytic Fungal Species. J. Bioremediation Biodegrad. 2017, 8.
- Oghenekaro, A.O.; Kovalchuk, A.; Raffaello, T.; Camarero, S.; Gressler, M.; Henrissat, B.; Lee, J.; Liu, M.; Martínez, A.T.; Miettinen, O.; et al. Genome sequencing of Rigidoporus microporus provides insights on genes important for wood decay, latex tolerance and interspecific fungal interactions. Sci. Rep. 2020, 10, 1–15.




