
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Andres Salcedo Jordan | + 1099 word(s) | 1099 | 2021-04-16 08:53:50 | | | |
| 2 | Lindsay Dong | Meta information modification | 1099 | 2021-06-11 03:55:20 | | |
Video Upload Options
Downy mildews affect important crops and cause severe losses in production worldwide. Accurate identification and monitoring of these plant pathogens, especially at early stages of the disease, is fundamental in achieving effective disease control. The rapid development of molecular methods for diagnosis has provided more specific, fast, reliable, sensitive, and portable alternatives for plant pathogen detection and quantification than traditional approaches.
1. Downy Mildew Pathogens
Downy mildew (DM) pathogens include several species of obligate oomycetes that can cause devastating damage to commercial [1], landscape [2], and natural ecosystem plants [3][4][5]. Species such as Plasmopara viticola [6], Pseudoperonospora cubensis [7], Pseudoperonospora humuli [8][9], Peronospora belbahrii [10], Plasmopara obducens [11], Peronospora tabacina [12] Peronospora effusa [13], Peronosclerospora philippinensis, and Sclerophthora rayssiae var. zeae [14] have resulted in significant losses due to downy mildew epidemics around the world. In some instances, the epidemics have been so severe that they have prompted historical shifts in crop production [15][16][17][18]. In addition to the aggressiveness of these pathogens, fungicide insensitivity further compounds losses attributed to disease [19][20][21][22]. Thus, research to improve diagnostics and management of downy mildew pathogens has become a priority for the scientific community in recent years [23][24][25][26].
2. How to Find Downy Mildew Pathogens
The diagnostics of downy mildew diseases has mainly relied upon direct observation of symptoms and signs using the naked eye or hand lenses and microscopes [27]. This is possible after observing their sexual (e.g., antheridia and oogonia) and asexual structures (e.g., sporangiophores, sporangia, and zoospores) (Figure 1) involved in survival and dispersion, and because many downy mildew pathogens produce distinctive foliar signs and symptoms when colonizing a host plant [2][28][29]. However, such methods fall short when detection in seed or planting material is needed [8][23], when symptoms and/or signs are not characteristic enough, resulting in misdiagnosis [30], or when the pathogen identity to species, pathotype, or clade level has disease management implications [31] (Figure 2).
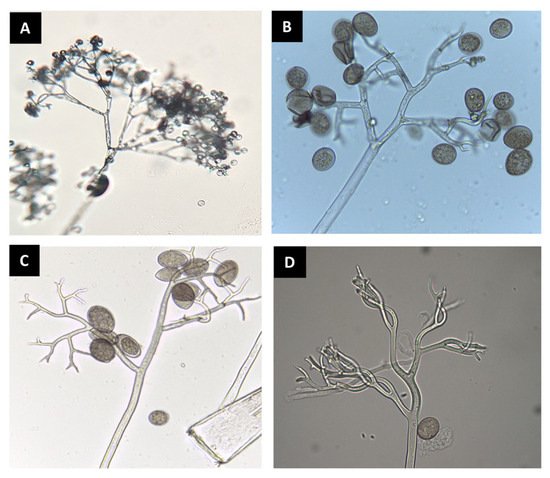
Figure 1. Sporangiophores and sporangia of downy mildew pathogens observed under a compound microscope. Bremia lactucae (A); Peronospora belbahrii (B); Pseudoperonospora cubensis (C); Peronospora chenopodii-ambrosioidis (D).
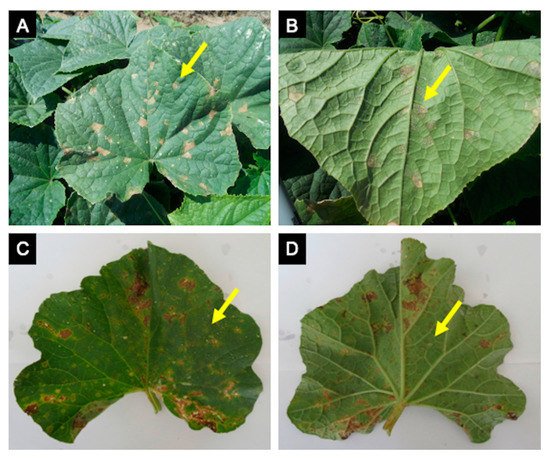
Figure 2. Cucurbit downy mildew caused by Pseudoperonospora cubensis in cucumber and cantaloupe. Cucumber symptoms (A) and signs (B) are very distinct, while cantaloupe symptoms (C) are often confused with other leaf spots or injury due to little sporulation on the underside of the leaf (D).
The rapid identification of disease-causing organisms as well as disease forecasting to reduce the use of pesticides [32][33] have created the necessity to develop more rapid, sensitive, versatile, high-throughput, and cost-efficient markers to identify and quantify plant pathogens. Unfortunately, downy mildew pathogens have been under-represented in oomycete phylogenetic studies and marker development [34]. Visual vs. molecular approaches for downy mildew diagnostics have different advantages and disadvantages [35], but rapid and accurate diagnostics are needed because, under favorable weather conditions, a field or greenhouse infected with downy mildew can result in complete loss of the crop in just a few days [36][37][38]. On-site visual inspection of symptoms and signs may provide a rapid diagnosis but requires trained personnel familiar with the particular downy mildew disease and the presence of distinct symptoms and/or signs [8][27][30]. Molecular diagnostic assays can be performed within a day but, in most cases, require a sample being taken to the laboratory, which can delay the process by several days [35]. This is also true when using conventional microscopy in the laboratory if on-site identification is not possible [39]. Molecular diagnostics can also provide additional information other than pathogen identity, such as fungicide sensitivity, virulence, aggressiveness, or pathogenicity, if such markers are available for a downy mildew pathogen [40][41]. However, for practical use of this information in disease management, results from assays need to be available quickly [39]. In this regard, field-deployable platforms for molecular diagnostics will be critical to unlock the potential for novel molecular markers and technologies to revolutionize disease management [42][43][44].
3. Different Marker Types
Visual-based diagnostic methods for plant pathogens are largely based on the observation of phenotypic characters and host range associations that require time and expertise for interpretation [45] and are frequently limited to taxonomic determination above species. By contrast, molecular techniques have changed the diagnostic emphasis towards pathogen presence and quantity using nucleic acids or proteins [46]. These techniques include serology (immunology), isoenzyme comparison, and nucleic acid-based technologies [44][46][47] (Table 1).
Table 1. Major advantages and disadvantages of molecular markers in the diagnosis of downy mildews.
| Source | Loci/Type | Advantages | Disadvantages | Examples |
|---|---|---|---|---|
| Nuclear | Ribosomal and Internal Transcribed Spacer (ITS) | -High reproducibility -Abundant copies -Common primers |
-Copy heterogeneity -Low resolution for cryptic species -Species cross-reactivity |
Pe destructor [48] Pe arborescens [49] Ps belbahrii [[50] Plasmopara spp. [51] B.lactucae [51] Ps humuli [52] |
| Housekeeping | -Common primers -known genes |
-Low polymorphisms and reproducibility -Limited for phylogenetic analysis |
Ps belbahrii [26] Ps cubensis [53] |
|
| Species-specific | -species-specific primers |
-Low polymorphisms and reproducibility -Limited for phylogenetic analysis |
Ps humuli [23] Ps cubensis [[40][41] |
|
| Multilocus | -Improves phylogenetic interpretation -Infraspecific resolution -High variability |
-Low reproducibility -More labor |
Ps cubensis [54] Ps. humuli [54]. |
|
| Mitochondrial | Single locus | -Improves phylogenetic interpretation -Infraspecific resolution -High variability |
-Uniparental heritance -Limited in detecting hybrid species |
B. lactucae [55] Ps cubensis [56] |
| Antigen | ELISA | -Speed and simplicity -High-throughput |
-Requires monoclonal antibodies -Species cross-reactivity -Limited use for biosurveillance |
Pe destructor [57] |
| Immunostrips | -Speed and simplicity -Cost-effective -Portability |
-Requires monoclonal antibody -Species cross-reactivity |
Still not available for downy mildew pathogens | |
| Enzymatic profile | Isozymes | -Codominant markers -Complement to phenotypic data |
-Large amount of tissue is required -Low polymorphisms -Influenced by the environment |
Pernosclerospora spp. [58] P. halstedii [59] |
The detection and identification of downy mildew pathogens are fundamental for disease management. Molecular diagnostic assays do not have most of the disadvantages associated with traditional detection methods, but the selection of a particular molecular assay is not free of caveats and depends on the main objective (detection, biosurveillance, pathogen relatedness, decision making on fungicide use, pathogen population structure, etc.). However, the development of more accurate DNA-based tests and better molecular markers for the detection of species and infraspecific taxonomic categories will continue as we increase the genetic and genomic information about downy mildew pathogens.
4. Future Prospects
While morphological features of downy mildew pathogens are identifiable by trained experts, using molecular inoculum detection tools represents a diagnostic tool to identify downy mildews with high specificity. The molecular tools available range from expensive technical formats with the ability to quantitate rapidly (e.g., qPCR) to handheld devices for in-field detection (e.g., LAMP, RPA), which give diverse options for finding downy mildews. Furthermore, given the potential for genetic differences informing management decisions (e.g., fungicide sensitivity of species or clades), molecular inoculum detection tools can be used for fungicide timing and selection by non-expert users. Ultimately, the goal of these approaches is to bring the diagnostic tool closer to the point of care with a tool that is simple, quick, and highly accurate that can detect and diagnose diseases. Future research in these areas should allow us to more accurately find these fantastic downy mildews.
References
- Gascuel, Q.; Martinez, Y.; Boniface, M.-C.; Vear, F.; Pichon, M.; Godiard, L. The sunflower downy. mildew pathogen Plasmopara halstedii: Plasmopara halstedii, sunflower downy mildew. Mol. Plant Pathol. 2015, 16, 109–122.
- Salgado-Salazar, C.; Shiskoff, N.; Daughtrey, M.; Palmer, C.L.; Crouch, J.A. Downy Mildew: A serious disease threat to rose health worldwide. Plant Dis. 2018, 102, 1873–1882.
- Wallace, E.C.; Quesada-Ocampo, L.M. First report of Downy Mildew on Buffalo Gourd (Cucurbita foetidissima) caused by Pseudoperonospora cubensis in North Carolina. Plant Dis. 2015, 99, 1861.
- Wallace, E.C.; Adams, M.; Ivors, K.; Ojiambo, P.S.; Quesada-Ocampo, L.M. First report of Pseudoperonospora cubensis causing Downy Mildew on Momordica balsamina and M. charantia in North Carolina. Plant Dis. 2014, 98, 1279.
- Wallace, E.; Choi, Y.J.; Thines, M.; Quesada-Ocampo, L.M. First Report of Plasmopara aff. australis on Luffa cylindrica in the United States. Plant Dis. 2015, 100, 537.
- Dussert, Y.; Mazet, I.D.; Couture, C.; Gouzy, J.; Piron, M.-C.; Kuchly, C.; Bouchez, O.; Rispe, C.; Mestre, P.; Delmotte, F. A High-Quality Grapevine Downy Mildew genome assembly reveals rapidly evolving and lineage-specific putative host adaptation genes. GBE 2019, 11, 954–969.
- Savory, E.A.; Granke, L.L.; Quesada-Ocampo, L.M.; Varbanova, M.; Hausbeck, M.K.; Day, B. The cucurbit downy mildew pathogen Pseudoperonospora cubensis. Mol. Plant Pathol. 2011, 12, 217–226.
- Purayannur, S.; Miles, T.D.; Gent, D.H.; Pigg, S.; Quesada-Ocampo, L.M. Hop Downy Mildew Caused by Pseudoperonospora humuli: A diagnostic guide. Plant Health Prog. 2020, 19, 173–179.
- Coley-Smith, J.R. Persistence and identification of downy mildew Pseudoperonospora humuli (Miy. and Tak.) Wilson in hop rootstocks. Ann. Appl. Biol. 1964, 53, 129–132.
- Wyenandt, C.A.; Simon, J.E.; Pyne, R.M.; Homa, K.; McGrath, M.T.; Zhang, S.; Raid, R.N.; Ma, L.-J.; Wick, R.; Guo, L.; et al. Basil Downy Mildew (Peronospora belbahrii): Discoveries and challenges relative to its control. Phytopathology 2015, 105, 885–894.
- Salgado-Salazar, C.; Rivera, Y.; Veltri, D.; Crouch, J.A. Polymorphic SSR markers for Plasmopara obducens (Peronosporaceae), the newly emergent downy mildew pathogen of Impatiens (Balsaminaceae). APPS 2015, 3, 1500073.
- Derevnina, L.; Chin-Wo-Reyes, S.; Martin, F.; Wood, K.; Froenicke, L.; Spring, O.; Michelmore, R. Genome sequence and architecture of the tobacco downy mildew pathogen Peronospora tabacina. MPMI 2015, 28, 1198–1215.
- Choi, Y.-J.; Hong, S.-B.; Shin, H.-D. Re-consideration of Peronospora farinosa infecting Spinacia oleracea as distinct species, Peronospora effusa. Mycolo. Res. 2007, 111, 381–391.
- Sharma, R.C.; De Leon, C.; Payak, M.M. Diseases of maize in South and South-East Asia: Problems and progress. Crop. Prot. 1993, 12, 414–422.
- Beans, C. Scientists Are Fighting for the Stricken Pickle against This Tricky Disease [Radio Broadcast]. NPR Radio. 14 December 2018. Available online: (accessed on 18 February 2021).
- Edwardson, J.R. Hops—Their botany, history, production and utilization. Econ. Bot. 1952, 6, 160–175.
- Holmes, G.J.; Ojiambo, P.S.; Hausbeck, M.K.; Quesada-Ocampo, L.M.; Keinath, A.P. Resurgence of cucurbit downy mildew in the United States: A watershed event for research and extension. Plant Dis. 2015, 99, 428–441.
- Frederiksen, R.A.; Renfro, B.L. Global status of maize downy mildew. Annu. Rev. Phytopathol. 1977, 15, 249–271.
- Chen, W.-J.; Delmotte, F.; Cervera, S.R.; Douence, L.; Greif, C.; Corio-Costet, M.-F. At least two origins of fungicide resistance in grapevine downy mildew populations. Appl. Environ. Microbiol. 2007, 73, 5162–5172.
- Nelson, M.E.; Eastwell, K.C.; Grove, G.G.; Barbour, J.D.; Ocamb, C.M.; Alldredge, J.R. Sensitivity of Pseudoperonospora humuli (the causal agent of hop downy mildew) from Oregon, Idaho, and Washington to fosetyl-Al (Aliette). Plant Health Prog. 2004, 5, 4.
- Pintore, I.; Gilardi, G.; Gullino, M.L.; Garibaldi, A. Detection of mefenoxam-resistant strains of Peronospora belbahrii, the causal agent of basil downy mildew, transmitted through infected seeds. Phytoparasitica 2016, 44, 563–569.
- Keinath, A.P.; Miller, S.A.; Smart, C.D. Response of Pseudoperonospora cubensis to preventative fungicide applications varies by state and year. Plant Health Prog. 2019, 20, 142–146.
- Rahman, A.; Góngora-Castillo, E.; Bowman, M.J.; Childs, K.L.; Gent, D.H.; Martin, F.N.; Quesada-Ocampo, L.M. Genome sequencing and transcriptome analysis of the hop downy mildew pathogen Pseudoperonospora humuli reveal species-specific genes for molecular detection. Phytopathology 2019, 109, 1354–1366.
- Withers, S.; Gongora-Castillo, E.; Gent, D.; Thomas, A.; Ojiambo, P.S.; Quesada-Ocampo, L.M. Using next-generation sequencing to develop molecular diagnostics for Pseudoperonospora cubensis, the cucurbit downy mildew pathogen. Phytopathology 2016, 106, 1105–1116.
- Blanco-Meneses, M.; Ristaino, J.B. Detection and quantification of Peronospora tabacina using a real-time polymerase chain reaction assay. Plant Dis. 2011, 95, 673–682.
- Shao, D.; Tian, M. A qPCR approach to quantify the growth of basil downy mildew pathogen Peronospora belbahrii during infection. Curr. Plant Biol. 2018, 15, 2–7.
- Salcedo, A.; Hausbeck, M.; Pigg, S.; Quesada-Ocampo, L.M. Diagnostic guide for cucurbit downy mildew. Plant Health Prog. 2020, 19, 166–172.
- Kandel, S.L.; Mou, B.; Shishkoff, N.; Shi, A.; Subbarao, K.V.; Klosterman, S.J. Spinach downy mildew: Advances in our understanding of the disease cycle and prospects for disease management. Plant Dis. 2019, 103, 791–803.
- Cohen, Y.; Van den Langenberg, K.M.; Wehner, T.C.; Ojiambo, P.S.; Hausbeck, M.K.; Quesada-Ocampo, L.M.; Lebeda, A.; Sierotzki, H.; Gisi, U. Resurgence of Pseudoperonospora cubensis: The causal agent of cucurbit downy mildew. Phytopathology 2015, 105, 998–1012.
- Standish, J.R.; Raid, R.N.; Pigg, S.; Quesada-Ocampo, L.M. A Diagnostic guide for basil downy mildew. Plant Health Prog. 2020, 21, 77–81.
- Wallace, E.C.; D’Arcangelo, K.N.; Quesada-Ocampo, L.M. Population analyses reveal two host-adapted clades of Pseudoperonospora cubensis, the causal agent of cucurbit downy mildew, on commercial and wild cucurbits. Phytopathology 2020, 110, 1578–1587.
- Martinelli, F.; Scalenghe, R.; Davino, S.; Panno, S.; Scuderi, G.; Ruisi, P.; Villa, P.; Stroppiana, D.; Boschetti, M.; Goulart, L.R.; et al. Advanced methods of plant disease detection. A review. Agron. Sustain. Dev. 2015, 35, 1–25.
- The Royal Society. Reaping the Benefits Science and the Sustainable Intensification of Global Agriculture; The Royal Society: London, UK, 2009.
- Klein, J. Genome reconstruction of the non-culturable spinach downy mildew Peronospora effusa by metagenome filtering. PLoS ONE 2020, 15, e0225808.
- Crandall, S.G.; Rahman, A.; Quesada-Ocampo, L.M.; Martin, F.N.; Bilodeau, G.J.; Miles, T.D. Advances in diagnostics of downy mildews: Lessons learned from other oomycetes and future challenges. Plant Dis. 2018, 102, 265–275.
- Aegerter, B.J.; Nuñez, J.J.; Davis, R.M. Environmental factors affecting rose downy mildew and development of a forecasting model for a nursery production system. Plant Dis. 2003, 87, 732–738.
- Granke, L.L.; Morrice, J.J.; Hausbeck, M.K. Relationships between airborne Pseudoperonospora cubensis sporangia, environmental conditions, and cucumber downy mildew severity. Plant Dis. 2014, 98, 674–681.
- Choudhury, R.A.; Koike, S.T.; Fox, A.D.; Anchieta, A.; Subbarao, K.V.; Klosterman, S.J.; McRoberts, N. Season-long dynamics of spinach downy mildew determined by spore trapping and disease incidence. Phytopathology 2016, 106, 1311–1318.
- Miller, S.A.; Beed, F.D.; Harmon, C.L. Plant disease diagnostic capabilities and networks. Annu. Rev. Phytopathol. 2009, 47, 15–38.
- Rahman, A.; Standish, J.; D’Arcangelo, K.N.; Quesada-Ocampo, L.M. Clade-specific biosurveillance of Pseudoperonospora cubensis using spore traps for precision disease management of cucurbit downy mildew. Phytopathology 2020, 6, 231.
- Rahman, A.; Miles, T.D.; Martin, F.N.; Quesada-Ocampo, L.M. Molecular approaches for biosurveillance of the cucurbit downy mildew pathogen, Pseudoperonospora cubensis. Can. J. Plant Pathol. 2017, 39, 282–296.
- Li, Z.; Paul, R.; Ba Tis, T.; Saville, A.C.; Hansel, J.C.; Yu, T.; Ristaino, J.B.; Wei, Q. Non-invasive plant disease diagnostics enabled by smartphone-based fingerprinting of leaf volatiles. Nat. Plants 2019, 5, 856–866.
- Marx, V. PCR heads into the field. Nat. Methods 2015, 12, 393–397.
- Kong, X.; Qin, W.; Huang, X.; Kong, F.; Schoen, C.D.; Feng, J.; Wang, Z.; Zhang, H. Development and application of loop-mediated isothermal amplification (LAMP) for detection of Plasmopara viticola. Sci. Rep. 2016, 6, 28935.
- Riethmuller, A.; Voglmayr, H.; Goker, M.; Weiss, M.; Oberwinkler, F. Phylogenetic relationships of the downy mildews (Peronosporales) and related groups based on nuclear large subunit ribosomal DNA sequences. Mycologia 2002, 94, 834.
- Goulter, K.; Randles, J. Serological and molecular techniques to detect and identify plant pathogens. In Plant Pathogens and Plant Diseases; Brown, J.F., Ogle, H.J., Eds.; Rockvale Publications: Armidale, Australia, 1997; pp. 172–191.
- Aslam, S.; Tahir, A.; Aslam, M.F.; Alam, M.W.; Shedayi, A.A.; Sadia, S. Recent advances in molecular techniques for the identification of phytopathogenic fungi–a mini review. J. Plant Interact. 2017, 12, 493–504.
- Van der Heyden, H.; Bilodeau, G.; Carisse, O.; Charron, J.-B. Monitoring of Peronospora destructor primary and secondary inoculum by real-time qPCR. Plant Dis. 2020, 104, 3183–3191.
- Montes-Borrego, M.; Muñoz-Ledesma, F.J.; Jiménez-Díaz, R.M.; Landa, B.B. Real-time PCR quantification of Peronospora arborescens, the opium poppy downy mildew pathogen, in seed stocks and symptomless infected plants. Plant Dis. 2011, 95, 143–152.
- Farahani-Kofoet, R.D.; Römer, P.; Grosch, R. Systemic spread of downy mildew in basil plants and detection of the pathogen in seed and plant samples. Mycol. Prog. 2012, 11, 961–966.
- Voglmayr, H.; Riethmüller, A.; Göker, M.; Weiss, M.; Oberwinkler, F. Phylogenetic relationships of Plasmopara, Bremia and other genera of downy mildew pathogens with pyriform haustoria based on Bayesian analysis of partial LSU rDNA sequence data. Mycol. Res. 2004, 108, 1011–1024.
- Gent, D.H.; Nelson, M.E.; Farnsworth, J.L.; Grove, G.G. PCR detection of Pseudoperonospora humuli in air samples from hop yards. Plant Pathol. 2009, 58, 1081–1091.
- Lee, J.H.; Park, M.H.; Lee, S. Identification of Pseudoperonospora cubensis using real-time PCR and high- resolution melting (HRM) analysis. J. Gen. Plant Pathol. 2016, 82, 110–115.
- Runge, F.; Choi, Y.-J.; Thines, M. Phylogenetic investigations in the genus Pseudoperonospora reveal overlooked species and cryptic diversity in the P. cubensis species cluster. Eur. J. Plant Pathol. 2011, 129, 135–146.
- Kunjeti, S.G.; Anchieta, A.; Martin, F.N.; Choi, Y.-J.; Thines, M.; Michelmore, R.W.; Koike, S.T.; Tsuchida, C.; Mahaffee, W.; Subbarao, K.V.; et al. Detection and quantification of Bremia lactucae by spore trapping and quantitative PCR. Phytopathology 2016, 106, 1426–1437.
- Summers, C.F.; Adair, N.L.; Gent, D.H.; McGrath, M.T.; Smart, C.D. Pseudoperonospora cubensis and P. humuli detection using species-specific probes and high-definition melt curve analysis. Can. J. Plant Pathol. 2015, 37, 315–330.
- Kennedy, R.; Wakeham, A.J. Development of detection systems for the sporangia of Peronospora destructor. In The Downy Mildews-Genetics, Molecular Biology and Control; Lebeda, A., Spencer-Phillips, P.T.N., Cooke, B.M., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 147–155.
- Bonde, M.R. Isozyme Analysis to differentiate species of Peronosclerospora causing downy mildews of maize. Phytopathology 1984, 74, 1278.
- Komjáti, H.; Bakonyi, J.; Spring, O.; Virányi, F. Isozyme analysis of Plasmopara halstedii using cellulose-acetate gel electrophoresis. Plant Pathol. 2007, 57, 57–63.




