Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Andrew Foers | + 2172 word(s) | 2172 | 2021-05-21 05:22:59 | | | |
| 2 | Bruce Ren | -21 word(s) | 2151 | 2021-05-24 03:30:28 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Foers, A. EV miRNAs Modulate Inflammation. Encyclopedia. Available online: https://encyclopedia.pub/entry/9937 (accessed on 07 February 2026).
Foers A. EV miRNAs Modulate Inflammation. Encyclopedia. Available at: https://encyclopedia.pub/entry/9937. Accessed February 07, 2026.
Foers, Andrew. "EV miRNAs Modulate Inflammation" Encyclopedia, https://encyclopedia.pub/entry/9937 (accessed February 07, 2026).
Foers, A. (2021, May 21). EV miRNAs Modulate Inflammation. In Encyclopedia. https://encyclopedia.pub/entry/9937
Foers, Andrew. "EV miRNAs Modulate Inflammation." Encyclopedia. Web. 21 May, 2021.
Copy Citation
In rheumatoid arthritis (RA), extracellular vesicles (EVs) are associated with both the propagation and attenuation of joint inflammation and destruction. However, the specific EV content responsible for these processes is largely unknown. Investigations into identifying EV content are confounded by the challenges in obtaining high-quality EV preparations from synovial fluid. Implementing a size exclusion chromatography-based method of EV isolation, coupled with small RNA sequencing, we accurately characterised EV miRNAs in synovial fluid obtained from RA patients and investigated the differences between joints with high- and low-grade inflammation.
rheumatoid arthritis
extracellular vesicles
miRNA
synovial fluid
1. Introduction
Rheumatoid arthritis (RA) is a chronic systemic autoimmune disease that targets synovial joints. RA causes chronic, progressive joint inflammation with exacerbations and remissions. Unless it is adequately treated, disease persistence leads to irreversible joint destruction and subsequent joint deformity, resulting in progressive disability. In RA, joint damage occurs due to an immune response that drives inflammatory cell infiltration. The pathophysiology of RA is incompletely understood. To improve patient outcomes and guide the development of more effective ways to control this disease, a better understanding of the disease processes is required.
Extracellular vesicles (EVs)—including exosomes, microvesicles and apoptotic bodies—are phospholipid bilayer-bound vesicles that are released from host cells and which contain a bioactive cargo of RNAs, proteins and lipids [1]. In RA, synovial fluid (SF) EVs have immunomodulatory functionality and are described to both contribute to and protect against joint destruction [2][3].
MicroRNAs (miRNAs) are ~18–25 nucleotide RNAs that can regulate protein translation through complementary binding to mRNA transcripts [4]. In RA, fluctuations in the expression of miRNAs may contribute to the severity of joint inflammation and destruction. For example, elevated miR-146a levels are reported in RA SF [5] and in CD4+ T cells isolated from RA SF [6], but not in osteoarthritis SF, whereas miR-146a expression in SF CD4+ T cells is positively correlated with SF TNF levels [6]. A direct role for miR-146a in promoting inflammation through upregulating T cell activity is supported by miR-146a overexpression, suppressing apoptosis in Jurkat T-like cells [6].
An increasing number of studies have also demonstrated that miRNAs encapsulated within EVs can promote inflammatory disease. For instance, miRNAs in Treg-derived EVs supressed T helper 1 cell proliferation and pro-inflammatory cytokine release [7]. Conversely, GU-rich motifs present in some EV miRNAs have been described to act as endolysosomal TLR8 (TLR7 in mice) agonists [8]. Consistent with this, EVs in the sera of HIV patients were found to contain GU-rich viral miRNAs that act as ligands for TLR8 and stimulate TNF release in recipient macrophages [9].
2. Characterisation of EV Isolation
EVs were first prepared by means of size exclusion chromatography (SEC) from SF obtained from a cohort of RA patients with either high- or low-grade inflammation (Table 1). To confirm that EV preparations were of satisfactory quality, EV enrichments were assessed by Western blotting for the canonical EV markers: HSP70, syntenin, FLOT1, Rab 27b, TSG101 and annexin 1 (Figure 1A). Transmission electron microscopy confirmed the presence of EVs with minimal apparent non-EV contaminating material (Figure 1B). These observations are consistent with our previous report detailing SEC as a method for obtaining high-quality EV enrichments from SF [10]. Previously, we demonstrated that RA patients with high-grade synovial inflammation have increased concentrations of SF EVs [11]. Consistent with this, we detected a 3.5-fold increase in the EV miRNA concentration per mL of SF in joints with high-grade inflammation (p-value = 0.0017; Figure 1C), suggesting the potential involvement of SF EV miRNAs in inflammatory processes in RA.
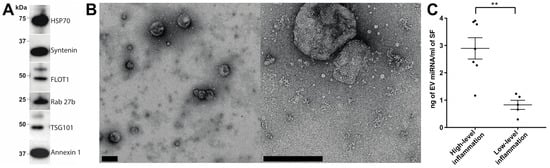
Figure 1. Assessment of synovial fluid EV enrichments prepared using size-exclusion chromatography. (A) Synovial fluid EV enrichments prepared by SEC were assessed for the presence of the canonical EV markers (HSP70, syntenin, FLOT1, Rab 27b, TSG101 and annexin 1) via Western blotting. (B) The quality of EV enrichments was further assessed by transmission electron microscopy (scale bars = 200 nm). (C) miRNA concentration per mL of rheumatoid arthritis synovial fluid in joints with high- or low-grade joint inflammation. Group means are indicated. Data analysed with Student’s i-test. Error bars = SEM. ** denotes p-value < 0.01.
Table 1. Summary of patient details and clinical parameters.
| RA (High-Grade Inflammation) | RA (Low-Grade Inflammation) | p-Value | |
|---|---|---|---|
| n | 7 | 5 | - |
| Age—mean (s.d.) | 64.4 (11.8) | 67.6 (10.0) | 0.64 |
| Sex—number of females/males | 3/4 | 2/3 | >0.99 |
| White cell count—mean (s.d.) cells µL−1 | 8940 * | 171.8 | <0.001 |
| Anti-citrullinated protein antibody (% positive) | 71% | 20% | 0.24 |
| Rheumatoid factor (% positive) | 86% | 60% | 0.52 |
| Disease Activity Score 28—median (range) | 4.7 (3.31–5.41) | 3.5 (2.74–5.0) | 0.15 |
| C-reactive protein—median (range) mg L−1 | 20 (6–164) | 2 (1.4–2) | 0.16 |
3. Highly Ranked SF EV miRNAs Target Immunomodulatory SF EV Proteins
To characterise the miRNA present in the SF of RA patients, small RNA sequencing was performed. A total of 1415 miRNA species were identified and, after filtering out lowly abundant species, 318 miRNAs remained. The 10 highest-ranked miRNAs across the entire patient cohort are specified in Table 2. To investigate biological processes associated with these prevalent miRNAs, experimentally validated gene targets were identified using miRTarBase and biological processes associated with the gene targets were characterised (Figure 2). A robust enrichment for genes involved in regulating gene expression was observed, including AKT1 (miR-100-5p, miR-10b-5p and miR-99a-5p), FOXO1 (let-7a-5p and miR-21-5p), KRAS (let-7a-5p), IKBKB (miR-148a-3p), STAT3 (let-7a-5p, miR-148a-3p, miR-21-5p and miR-92a-3p), TGFBR1 (let-7b-5p) and TP53 (miR-10b-5p). In addition, genes associated with cytokine-mediated signalling were targets of highly prevalent miRNA, including the inflammatory mediators CCR1 (miR-21-5p), IL-1β (miR-21-5p), IL-6 (let-7a-5p and miR-26a-5p), MCL1 (miR-26a-5p), NOS2 (miR-26a-5p), PIK3CA (miR-10b-5p) and SOCS5 (miR-92a-3p).
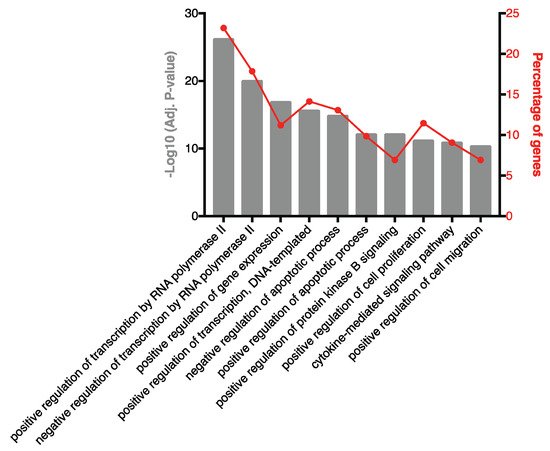
Figure 2. Biological processes associated with experimentally validated targets of highly ranked SF EV miRNA. Pathway analysis was performed on experimentally validated gene targets of the 10 highest-ranked miRNAs. The percentage of target genes associated with each biological process are indicated.
Table 2. Highest-ranked SF EV miRNAs across the entire patient cohort.
| Rank | miRNA | Average Expression (CPM) |
|---|---|---|
| 1 | hsa-miR-100-5p | 132,441 |
| 2 | hsa-miR-21-5p | 179,929 |
| 3 | hsa-miR-148a-3p | 86,059 |
| 4 | hsa-let-7a-5p | 43,627 |
| 5 | hsa-miR-92a-3p | 45,881 |
| 6 | hsa-let-7b-5p | 31,280 |
| 7 | hsa-miR-10b-5p | 35,499 |
| 8 | hsa-miR-99b-5p | 31,569 |
| 9 | hsa-miR-26a-5p | 34,975 |
| 10 | hsa-miR-99a-5p | 45,879 |
To investigate if these 10 highly-ranked miRNAs might synergise to repress the expression of individual genes, common gene targets were identified. Genes targeted by four or more miRNA are listed in Table 3. Interestingly, the Akt activator insulin-like growth factor 1 receptor (IGF1R) is a target of five of the 10 highest-ranked miRNAs, and the pro-inflammatory transcription factor STAT3 is targeted by four of the 10 highest-ranked miRNAs.
Table 3. Common targets of the 10 highest-ranked EV miRNAs.
| arget Gene ID | Target Gene Name | miRNA Regulators |
|---|---|---|
| IGF1R | Insulin like growth factor 1 receptor | hsa-let-7b-5p hsa-miR-100-5p hsa-miR-21-5p hsa-miR-99a-5p hsa-miR-99b-5p |
| CCND2 | Cyclin D2 | hsa-let-7a-3p hsa-let-7a-5p hsa-let-7b-5p hsa-miR-26a-5p |
| E2F2 | E2F transcription factor 2 | hsa-let-7a-3p hsa-let-7a-5p hsa-let-7b-5p hsa-miR-26a-5p |
| PTEN | Phosphatase and tensin homolog | hsa-miR-10b-5p hsa-miR-21-5p hsa-miR-26a-5p hsa-miR-92a-3p |
| STAT3 | Signal transducer and activator of transcription 3 | hsa-let-7a-5p hsa-miR-148a-3p hsa-miR-21-5p hsa-miR-92a-3p |
4. Seventy-Eight SF EV miRNAs Are Differentially Expressed between RA Patients with High- and Low-Grade Inflammation
To investigate how SF EV miRNAs might more specifically contribute to RA pathophysiology, differences in miRNA expression in RA joints with either high- or low-grade inflammation were characterised. Overall, 78 differentially expressed miRNAs were defined. Of these, 49 were elevated in high-grade inflammation, whereas 29 were elevated in RA joints with low-grade inflammation (Figure 3A and Table 4).
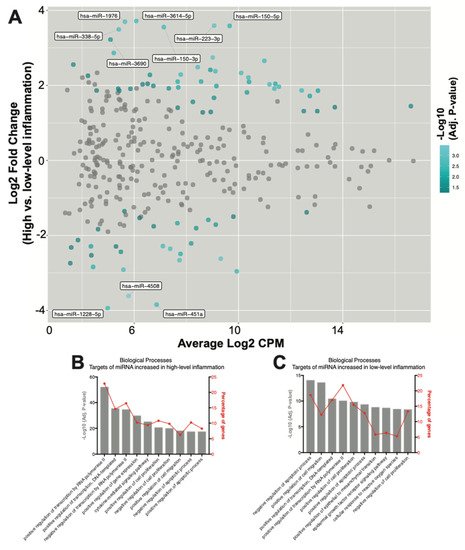
Figure 3. SF EV miRNAs have diverse immunoregulatory capacities. (A) MA plot of miRNA counts per million reads (CPM) vs. fold change. Differentially expressed miRNAs with an adjusted p-value < 0.05 are highlighted in blue. miRNAs with an adjusted p-value < 0.05 and a log2 fold change >3 are labelled. (B,C) Biological processes associated with target genes of the miRNAs significantly increased in joints with (B) high-, and (C) low-grade inflammation. The percentage of target genes associated with each biological process is indicated.
Table 4. Differentially expressed miRNAs.
| miRNA | Average Read Count (Log2 CPM) | Log2 Fold Change(High- vs. Low−Grade Inflammation) | Adjusted p-Value |
|---|---|---|---|
| hsa-miR-4508 | 5.78 | −3.61 | 3.94 × 10−4 |
| hsa-miR-223-3p | 9.07 | 3.59 | 6.42 × 10−4 |
| hsa-miR-3529-3p | 9.01 | 2.74 | 6.42 × 10−4 |
| hsa-miR-615-3p | 9.32 | −2.62 | 6.42 × 10−4 |
| hsa-miR-1976 | 5.65 | 3.70 | 1.15 × 10−3 |
| hsa-miR-543 | 7.74 | −2.66 | 1.15 × 10−3 |
| hsa-miR-338-5p | 5.41 | 3.49 | 1.36 × 10−3 |
| hsa-miR-146b-3p | 8.43 | 2.49 | 1.36 × 10−3 |
| hsa-miR-433-3p | 7.38 | −2.36 | 1.36 × 10−3 |
| hsa-miR-485-3p | 5.59 | −2.91 | 1.36 × 10−3 |
| hsa-miR-101-3p | 10.10 | 2.33 | 1.52 × 10−3 |
| hsa-miR-27a-5p | 11.19 | 2.03 | 1.53 × 10−3 |
| hsa-miR-361-3p | 11.38 | 1.91 | 1.76 × 10−3 |
| hsa-miR-3614-5p | 6.08 | 3.72 | 1.86 × 10−3 |
| hsa-miR-150-3p | 7.14 | 3.55 | 1.89 × 10−3 |
| hsa-miR-223-5p | 9.11 | 2.38 | 1.89 × 10−3 |
| hsa-miR-142-5p | 7.78 | 2.29 | 1.89 × 10−3 |
| hsa-miR-106b-3p | 10.38 | 2.21 | 1.89 × 10−3 |
| hsa-miR-28-3p | 12.42 | 1.87 | 1.89 × 10−3 |
| hsa-miR-455-5p | 7.10 | −2.28 | 1.89 × 10−3 |
| hsa-miR-451a | 6.86 | −3.84 | 1.89 × 10−3 |
| hsa-miR-143-3p | 10.99 | 2.66 | 2.04 × 10−3 |
| hsa-miR-1228-5p | 4.99 | −3.94 | 2.11 × 10−3 |
| hsa-miR-30e-3p | 11.55 | 1.96 | 2.25 × 10−3 |
| hsa-miR-486-5p | 9.93 | −2.96 | 2.30 × 10−3 |
| hsa-miR-1273h-3p | 5.22 | 2.87 | 2.56 × 10−3 |
| hsa-miR-150-5p | 9.66 | 3.59 | 3.01 × 10−3 |
| hsa-miR-378c | 7.67 | 2.10 | 3.61 × 10−3 |
| hsa-miR-92b-5p | 7.78 | −2.49 | 3.61 × 10−3 |
| hsa-miR-4448 | 4.42 | −2.84 | 3.61 × 10−3 |
| hsa-miR-103b | 8.57 | 2.10 | 4.76 × 10−3 |
| hsa-miR-941 | 11.41 | 2.04 | 4.76 × 10−3 |
| hsa-miR-103a-3p | 7.69 | 1.95 | 5.24 × 10−3 |
| hsa-miR-1246 | 10.40 | 2.09 | 5.64 × 10−3 |
| hsa-miR-125b-1-3p | 7.66 | −1.67 | 5.64 × 10−3 |
| hsa-miR-769-5p | 9.15 | 1.98 | 6.68 × 10−3 |
| hsa-miR-378a-3p | 13.05 | 1.86 | 6.68 × 10−3 |
| hsa-miR-140-3p | 12.73 | 1.72 | 6.68 × 10−3 |
| hsa-miR-214-5p | 4.34 | −2.29 | 6.68 × 10−3 |
| hsa-miR-574-3p | 8.84 | −1.59 | 7.83 × 10−3 |
| hsa-miR-1180-3p | 8.18 | −2.21 | 8.45 × 10−3 |
| hsa-miR-155-5p | 10.33 | 1.81 | 8.69 × 10−3 |
| hsa-miR-629-5p | 8.92 | 2.41 | 9.41 × 10−3 |
| hsa-miR-328-3p | 9.13 | −1.71 | 1.05 × 10−2 |
| hsa-miR-3690 | 5.09 | 3.22 | 1.18 × 10−2 |
| hsa-miR-7704 | 8.38 | −1.81 | 1.48 × 10−2 |
| hsa-miR-221-5p | 9.06 | 1.67 | 1.51 × 10−2 |
| hsa-miR-486-3p | 5.49 | −2.44 | 1.51 × 10−2 |
| hsa-miR-589-5p | 6.60 | 2.28 | 1.56 × 10−2 |
| hsa-miR-142-3p | 7.33 | 1.92 | 1.56 × 10−2 |
| hsa-miR-192-5p | 8.25 | 1.56 | 1.98 × 10−2 |
| hsa-miR-618 | 3.67 | 2.55 | 2.18 × 10−2 |
| hsa-miR-21-5p | 16.61 | 1.45 | 2.22 × 10−2 |
| hsa-miR-21-3p | 6.73 | 1.99 | 2.57 × 10−2 |
| hsa-miR-345-5p | 5.86 | 1.91 | 2.59 × 10−2 |
| hsa-miR-214-3p | 6.75 | −1.76 | 2.59 × 10−2 |
| hsa-miR-532-5p | 12.50 | 1.31 | 2.70 × 10−2 |
| hsa-miR-378e | 5.43 | 1.86 | 2.86 × 10−2 |
| hsa-miR-185-3p | 6.32 | 2.03 | 3.05 × 10−2 |
| hsa-miR-6787-3p | 3.55 | −2.74 | 3.07 × 10−2 |
| hsa-miR-503-5p | 4.23 | 2.26 | 3.11 × 10−2 |
| hsa-miR-25-3p | 13.02 | 1.31 | 3.11 × 10−2 |
| hsa-miR-3120-5p | 4.61 | −1.83 | 3.24 × 10−2 |
| hsa-miR-203b-5p | 3.84 | −2.12 | 3.35 × 10−2 |
| hsa-miR-500a-3p | 7.67 | 1.40 | 3.42 × 10−2 |
| hsa-miR-7-5p | 4.56 | 1.86 | 3.56 × 10−2 |
| hsa-miR-3622a-5p | 4.36 | −2.53 | 3.56 × 10−2 |
| hsa-miR-365a-5p | 5.30 | −2.29 | 3.76 × 10−2 |
| hsa-miR-23b-3p | 9.80 | −1.50 | 3.85 × 10−2 |
| hsa-miR-424-3p | 6.52 | 2.04 | 4.29 × 10−2 |
| hsa-miR-501-3p | 8.94 | 1.29 | 4.30 × 10−2 |
| hsa-miR-92b-3p | 12.61 | −1.38 | 4.37 × 10−2 |
| hsa-miR-146b-5p | 13.69 | 1.42 | 4.42 × 10−2 |
| hsa-miR-185-5p | 8.63 | 1.28 | 4.48 × 10−2 |
| hsa-miR-23b-5p | 6.21 | −1.53 | 4.58 × 10−2 |
| hsa-miR-378f | 5.39 | 1.91 | 4.73 × 10−2 |
| hsa-miR-27b-5p | 5.63 | −1.36 | 4.76 × 10−2 |
| hsa-miR-1306-5p | 3.58 | −2.32 | 4.98 × 10−2 |
To further investigate miRNA contributions within joints with high-grade inflammation, gene targets of the 49 miRNAs that increased in highly inflamed joints were identified and the associated biological processes were explored. Pathway analysis again revealed an enrichment for genes associated with processes regulating gene expression (Figure 3B). ‘Positive regulation of transcription by RNA polymerase II’ was the highest-ranked biological process, with 22% of all target genes associated. The ‘cytokine mediated signalling pathway’ was also again highly ranked, with target genes including AKT1 (miR-143-3p, miR-185-3p and miR-192-5p), CCL20 (miR-21-5p), CCL3 (miR-223-3p), CCR1 (miR-21-5p), CISH (miR-150-5p), CSF1R (miR-155-5p), CXCL8 (miR-155-5p), FASLG (miR-21-3p and miR-21-5p), IL-1β (miR-21-5p), IL6R (miR-221-5p), JAK2 (miR-101-3p), MMP9 (miR-143-3p), STAT1 (miR-150-5p, miR-155-5p and miR-223-3p), STAT3 (miR-21-3p, miR-21-5p and miR-223-3p), TGFB1 (miR-185-5p and miR-21-5p), TNF (miR-143-3p) and TP53 (miR-150-3p, miR-150-5p, miR-25-3p and miR-28-3p) (Table S7). Some of the 49 miRNAs found to be increased in EVs from highly inflamed joints may synergistically repress the translation of genes involved in driving inflammatory processes, as demonstrated by multiple miRNAs targeting proteins typically associated with promoting inflammation, including IGF1R, PTEN, VEGFA and BCL2 (Table 5).
Table 5. Common targets of SF EV miRNAs found to be significantly enriched in the joints of RA patients with high-grade inflammation.
| Target Gene ID | Target Gene Name | miRNA Regulators |
|---|---|---|
| IGF1R | Insulin like growth factor 1 receptor | hsa-miR-143-3p hsa-miR-150-3p hsa-miR-185-5p hsa-miR-21-5p hsa-miR-223-3p hsa-miR-223-5p hsa-miR-378a-3p hsa-miR-503-5p hsa-miR-7-5p |
| PTEN | Phosphatase and tensin homolog | hsa-miR-103a-3p hsa-miR-106b-3p hsa-miR-142-5p hsa-miR-155-5p hsa-miR-21-3p hsa-miR-21-5p hsa-miR-25-3p |
| VEGFA | Vascular endothelial growth factor A | hsa-miR-101-3p hsa-miR-150-5p hsa-miR-185-5p hsa-miR-21-5p hsa-miR-378a-3p hsa-miR-503-5p |
| BCL2 | BCL2, apoptosis regulator | hsa-miR-143-3p hsa-miR-192-5p hsa-miR-21-5p hsa-miR-503-5p hsa-miR-7-5p |
| FBXW7 | F-box and WD repeat domain containing 7 | hsa-miR-155-5p hsa-miR-223-3p hsa-miR-223-5p hsa-miR-25-3p hsa-miR-503-5p |
| MYB | MYB proto-oncogene, transcription factor | hsa-miR-103a-3p hsa-miR-150-3p hsa-miR-150-5p hsa-miR-155-5p hsa-miR-503-5p |
| EGFR | Epidermal growth factor receptor | hsa-miR-146b-5p hsa-miR-21-5p hsa-miR-27a-5p hsa-miR-7-5p |
| RAC1 | Rac family small GTPase 1 | hsa-miR-101-3p hsa-miR-142-3p hsa-miR-142-5p hsa-miR-155-5p |
| TP53 | Tumor protein p53 | hsa-miR-150-3p hsa-miR-150-5p hsa-miR-25-3p hsa-miR-28-3p |
| ZEB1 | Zinc finger E-box binding homeobox 1 | hsa-miR-101-3p hsa-miR-142-5p hsa-miR-150-5p hsa-miR-223-3p |
Finally, the functionality of the 29 miRNAs found to be increased in EVs within the joints of RA patients with low-grade inflammation were investigated. Pathway analysis of gene targets revealed an enrichment for genes associated with apoptotic processes, cell migration, gene expression and cell proliferation (Figure 3C). ‘Negative regulation of apoptotic processes’ was the highest-ranked biological process, with miRNA target genes including AKT1 (miR-451a), CD44 (miR-328-3p), IGF1R (miR-214-3p, miR-23b-3p, miR-486-5p and miR-92b-3p), IKBKB (miR-451a), IL6 (miR-451a), MIF (miR-1228-5p and miR-451a) and MYC (miR-451a). Interestingly, similarly to the above observations in joints with high-grade inflammation, multiple miRNAs found to be enriched in joints with low-grade inflammation again target PTEN and TP53 (Table 6).
Table 6. Common targets of SF EV miRNAs found to be significantly enriched in the joints of RA patients with low-grade inflammation.
| Target Gene ID | Target Gene Name | miRNA Regulators |
|---|---|---|
| PTEN | Phosphatase and tensin homolog | hsa-miR-214-3p hsa-miR-23b-3p hsa-miR-486-5p hsa-miR-92b-3p |
| TP53 | Tumor protein p53 | hsa-miR-125b-1-3p hsa-miR-214-3p hsa-miR-214-5p |
References
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659.
- Foers, A.D.; Cheng, L.; Hill, A.F.; Wicks, I.P.; Pang, K.C. Review: Extracellular Vesicles in Joint Inflammation. Arthritis Rheumatol. 2017, 69, 1350–1362.
- Malda, J.; Boere, J.; Van De Lest, C.H.A.; Van Weeren, P.R.; Wauben, M.H.M. Extracellular vesicles—New tool for joint repair and regeneration. Nat. Rev. Rheumatol. 2016, 12, 243–249.
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402.
- Murata, K.; Yoshitomi, H.; Tanida, S.; Ishikawa, M.; Nishitani, K.; Ito, H.; Nakamura, T. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res. Ther. 2010, 12, 1–14.
- Li, J.; Wan, Y.; Guo, Q.; Zou, L.; Zhang, J.; Fang, Y.; Zhang, J.; Zhang, J.; Fu, X.; Liu, H.; et al. Altered microRNA expression profile with miR-146a upregulation in CD4+ T cells from patients with rheumatoid arthritis. Arthritis Res. Ther. 2010, 12, R81.
- Okoye, I.S.; Coomes, S.M.; Pelly, V.S.; Czieso, S.; Papayannopoulos, V.; Tolmachova, T.; Seabra, M.C.; Wilson, M.S. MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity 2014, 41, 89–103.
- Bosch, S.; Young, N.A.; Mignot, G.; Bach, J.-M. Epigenetic Mechanisms in Immune Disease: The Significance of Toll-Like Receptor-Binding Extracellular Vesicle-Encapsulated microRNA. Front. Genet. 2020, 11, 578335.
- Bernard, M.A.; Zhao, H.; Yue, S.C.; Anandaiah, A.; Koziel, H.; Tachado, S.D. Novel HIV-1 MiRNAs Stimulate TNFα Release in Human Macrophages via TLR8 Signaling Pathway. PLoS ONE 2014, 9, e106006.
- Foers, A.D.; Chatfield, S.; Dagley, L.F.; Scicluna, B.J.; Webb, A.I.; Cheng, L.; Hill, A.F.; Wicks, I.P.; Pang, K.C. Enrichment of extracellular vesicles from human synovial fluid using size exclusion chromatography. J. Extracell. Vesicles 2018, 7, 1490145.
- Foers, A.D.; Dagley, L.F.; Chatfield, S.; Webb, A.I.; Cheng, L.; Hill, A.F.; Wicks, I.P.; Pang, K.C. Proteomic analysis of extracellular vesicles reveals an immunogenic cargo in rheumatoid arthritis synovial fluid. Clin. Transl. Immunol. 2020, 9, e1185.
More
Information
Subjects:
Immunology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
578
Revisions:
2 times
(View History)
Update Date:
24 May 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




