
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | simone sampaolo | + 2897 word(s) | 2897 | 2021-08-25 06:09:56 | | | |
| 2 | Peter Tang | Meta information modification | 2897 | 2021-09-01 04:18:12 | | |
Video Upload Options
Autophagy is the major intracellular machinery for degrading proteins, lipids, polysaccharides, and organelles. This cellular process is essential for the maintenance of the correct cellular balance in both physiological and stress conditions. Because of its role in maintaining cellular homeostasis, dysregulation of autophagy leads to various disease manifestations, such as inflammation, metabolic alterations, aging, and neurodegeneration. A common feature of many neurologic and neuromuscular diseases is the alteration of the autophagy-lysosomal pathways. For this reason, autophagy is considered a target for the prevention and/or cure of these diseases. Dietary intake of polyphenols has been demonstrated to prevent/ameliorate several of these diseases. Thus, natural products that can modulate the autophagy machinery are considered a promising therapeutic strategy. In particular, curcumin, a phenolic compound widely used as a dietary supplement, exerts an important effect in modulating autophagy.
1. Introduction
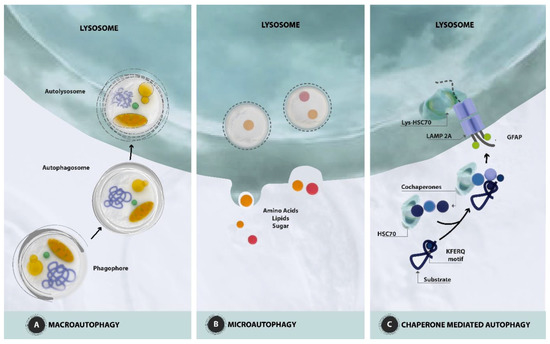
2. Autophagy: Mechanisms and Regulation
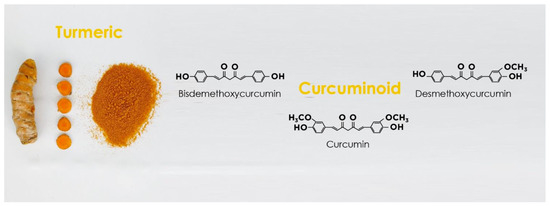
3. Curcumin Structure and Activity
4. Autophagy Modulation and the Interplay between Autophagy and Curcumin as a Therapeutic Approach for Neurological Disorders
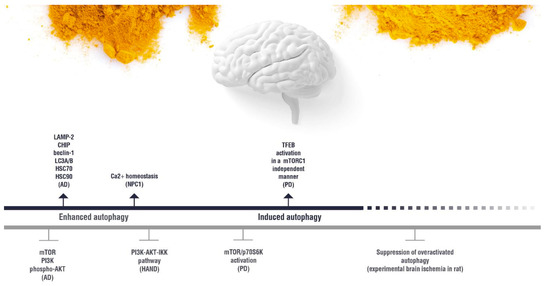
 (induction);
(induction);  (inhibition). AD: Alzheimer’s disease; HAND: HIV-induced neurocognitive disorder; PD: Parkinson’s disease; NPC1: Niemann Pick C1.
(inhibition). AD: Alzheimer’s disease; HAND: HIV-induced neurocognitive disorder; PD: Parkinson’s disease; NPC1: Niemann Pick C1.5. Effect of Curcumin on the Autophagy Pathways in Neuromuscular Diseases
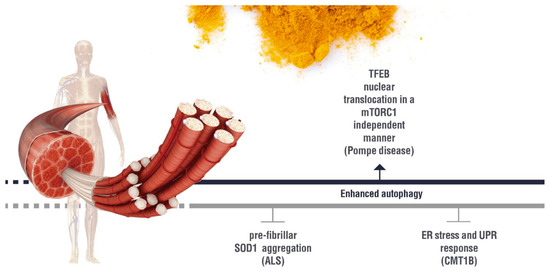
 (induction);
(induction);  (inhibition). ALS: Amyotrophic lateral sclerosis; ER:Endoplasmic Reticulum; UPR: Unfolded Protein Response; CMT1B: Charcot Marie Tooth 1B.
(inhibition). ALS: Amyotrophic lateral sclerosis; ER:Endoplasmic Reticulum; UPR: Unfolded Protein Response; CMT1B: Charcot Marie Tooth 1B.References
- Goel, A.; Kunnumakkara, A.B.; Aggarwal, B.B. Curcumin as “Curecumin”: From kitchen to clinic. Biochem. Pharmacol. 2008, 75, 787–809.
- Hosseini, A.; Hosseinzadeh, H. Antidotal or protective effects of Curcuma longa (turmeric) and its active ingredient, curcumin, against natural and chemical toxicities: A review. Biomed. Pharmacother. 2018, 99, 411–421.
- Ringman, J.M.; Frautschy, S.A.; Cole, G.M.; Masterman, D.L.; Cummings, J.L. A potential role of the curry spice curcumin in Alzheimer’s disease. Curr. Alzheimer Res. 2005, 2, 131–136.
- Shakeri, A.; Cicero, A.; Panahi, Y.; Sahebkar, A. Curcumin: A naturally occurring autophagy modulator. J. Cell Physiol. 2019, 234, 5643–5654.
- Soleimani, V.; Sahebkar, A.; Hosseinzadeh, H. Turmeric (Curcuma longa) and its major constituent (curcumin) as nontoxic and safe substances: Review. Phytother.Res. 2018, 32, 985–995.
- Aggarwal, B.; Sung, B. Pharmacological basis for the role of curcumin in chronic diseases: An age-old spice with modern targets. Trends Pharmacol. Sci. 2009, 30, 85–94.
- Vidoni, C.; Castiglioni, A.; Seca, C.; Secomandi, E.; Melone, M.; Isidoro, C. Dopamine exacerbates mutant Huntingtin toxicity via oxidative-mediated inhibition of autophagy in SH-SY5Y neuroblastoma cells: Beneficial effects of anti-oxidant therapeutics. Neurochem. Int. 2016, 101, 132–143.
- Vidoni, C.; Secomandi, E.; Castiglioni, A.; Melone, M.; Isidoro, C. Resveratrol protects neuronal-like cells expressing mutant Huntingtin from dopamine toxicity by rescuing ATG4-mediated autophagosome formation. Neurochem. Int. 2018, 117, 174–187.
- Galluzzi, L.; Baehrecke, E.; Ballabio, A.; Boya, P.; Bravo, S.P.J.; Cecconi, F.; Choi, A.; Chu, C.; Codogno, P.; Colombo, M. Molecular definitions of autophagy and related processes. EMBO J. 2017, 36, 1811–1836.
- Guo, S.; Long, M.; Li, X.; Zhu, S.; Zhang, M.; Yang, Z. Curcumin activates autophagy and attenuates oxidative damage in EA.hy926 cells via the Akt/mTOR pathway. Mol.Med.Rep. 2016, 13, 2187–2193.
- Salehi, B.; Stojanović, R.Z.; Matejić, J.; Sharifi, R.M.; Anil, K.N.; Martins, N.; Sharifi, R.J. The therapeutic potential of curcumin: A review of clinical trials. Eur.J.Med. Chem. 2019, 163, 527–545.
- Sridhar, S.; Botbol, Y.; Macian, F.; Cuervo, A. Autophagy and disease: Always two sides to a problem. J. Pathol. 2012, 226, 255–273.
- Lin, S.; Tsai, M.; Cheng, H.; Weng, C. Natural Compounds from Herbs that can Potentially Execute as Autophagy Inducers for Cancer Therapy. Int. J. Mol. Sci. 2017, 18, 1412.
- Bielak-Zmijewska, A.; Grabowska, W.; Ciolko, A.; Bojko, A.; Mosieniak, G.; Bijoch, Ł.; Sikora, E. The Role of Curcumin in the Modulation of Ageing. Int. J. Mol. Sci. 2019, 20, 1239.
- Guo, F.; Liu, X.; Cai, H.; Le, W. Autophagy in neurodegenerative diseases: Pathogenesis and therapy. Brain Pathol. 2018, 28, 3–13.
- Kondo, Y.; Kanzawa, T.; Sawaya, R.; Kondo, S. The role of autophagy in cancer development and response to therapy. Nat.Rev. Cancer 2005, 5, 726–734.
- Baehrecke, E.H. Autophagy: Dual roles in life and death? Nat.Rev.Mol. Cell Biol. 2005, 6, 505–510.
- Edinger, A.; Thompson, C. Death by design: Apoptosis, necrosis and autophagy. Curr. Opin. Cell Biol. 2004, 16, 663–669.
- Puri, C.; Renna, M.; Bento, C.; Moreau, K.; Rubinsztein, D. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 2013, 154, 1285–1299.
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42.
- Ohsumi, Y.; Mizushima, N. Two ubiquitin-like conjugation systems essential for autophagy. Semin. Cell Dev. Biol. 2004, 15, 231–236.
- Jung, C.; Jun, C.; Ro, S.; Kim, Y.; Otto, N.; Cao, J.; Kundu, M.; Kim, D. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell. 2009, 20, 1992–2003.
- Fan, W.; Nassiri, A.; Zhong, Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc. Natl. Acad. Sci. USA 2011, 108, 7769–7774.
- He, C.; Levine, B. The Beclin 1 interactome. Curr. Opin. Cell Biol. 2010, 22, 140–149.
- Shao, Y.; Gao, Z.; Feldman, T.; Jiang, X. Stimulation of ATG12-ATG5 conjugation by ribonucleic acid. Autophagy 2007, 3, 10–16.
- Fujita, N.; Hayashi-Nishino, M.; Fukumoto, H.; Omori, H.; Yamamoto, A.; Noda, T.; Yoshimori, T. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell. 2008, 19, 4651–4659.
- Kabeya, Y.; Mizushima, N.; Ueno, T.; Yamamoto, A.; Kirisako, T.; Noda, T.; Kominami, E.; Ohsumi, Y.; Yoshimori, T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000, 19, 5720–5728.
- Ravikumar, B.; Acevedo-Arozena, A.; Imarisio, S.; Berger, Z.; Vacher, C.; O’Kane, C.; Brown, S.; Rubinsztein, D. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat.Genet. 2005, 37, 771–776.
- Itakura, E.; Kishi-Itakura, C.; Mizushima, N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 2012, 151, 1256–1269.
- Cuervo, A.M. Autophagy: In sickness and in health. Trends Cell Biol. 2004, 14, 70–77.
- Meléndez, A.; Tallóczy, Z.; Seaman, M.; Eskelinen, E.; Hall, D.; Levine, B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 2003, 301, 1387–1391.
- Hong, C.J.; Park, H.; Yu, S.W. Autophagy for the quality control of adult hippocampal neural stem cells. Brain Res. 2016, 1649, 166–172.
- Scherz, S.R.; Elazar, Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007, 17, 422–427.
- Codogno, P.; Meijer, A.J. Autophagy and signaling: Their role in cell survival and cell death. Cell Death Differ. 2005, 12, 1509–1518.
- Sardiello, M.; Palmieri, M.; di Ronza, A.; Medina, D.; Valenza, M.; Gennarino, V.; di Malta, C.; Donaudy, F.; Embrione, V.; Polishchuk, R. A gene network regulating lysosomal biogenesis and function. Science 2009, 325, 473–477.
- Settembre, C.; Di Malta, C.; Polito, V.A.; Garcia Arencibia, M.; Vetrini, F.; Erdin, S.; Erdin, S.U.; Huynh, T.; Medina, D.; Colella, P.; et al. TFEB links autophagy to lysosomal biogenesis. Science 2011, 332, 1429–1433.
- Perrone, L.; Devi, T.S.; Hosoya, K.; Terasaki, T.; Singh, L.P. Thioredoxin interacting protein (TXNIP) induces inflammation through chromatin modification in retinal capillary endothelial cells under diabetic conditions. J. Cell Physiol. 2009, 221, 262–272.
- Perrone, L.; Devi, T.S.; Hosoya, K.I.; Terasaki, T.; Singh, L.P. Inhibition of TXNIP expression in vivo blocks early pathologies of diabetic retinopathy. Cell Death Dis. 2010, 1, e65.
- Perrone, L.; Sbai, O.; Nawroth, P.P.; Bierhaus, A. The Complexity of Sporadic Alzheimer’s Disease Pathogenesis: The Role of RAGE as Therapeutic Target to Promote Neuroprotection by Inhibiting Neurovascular Dysfunction. Int.J. Alzheimers Dis. 2012, 2012, 734956.
- Huang, C.; Zhang, Y.; Kelly, D.J.; Tan, C.Y.; Gill, A.; Cheng, D.; Braet, F.; Park, J.S.; Sue, C.; Pollock, C.A.; et al. Thioredoxin interacting protein (TXNIP) regulates tubular autophagy and mitophagy in diabetic nephropathy through the mTOR signaling pathway. Sci. Rep. 2016, 6, 29196.
- Qiao, S.; Dennis, M.; Song, X.; Vadysirisack, D.D.; Salunke, D.; Nash, Z.; Yang, Z.; Liesa, M.; Yoshioka, J.; Matsuzawa, S.; et al. A REDD1/TXNIP pro-oxidant complex regulates ATG4B activity to control stress-induced autophagy and sustain exercise capacity. Nat. Commun. 2015, 6, 7014.
- Crighton, D.; Wilkinson, S.; O’Prey, J.; Syed, N.; Smith, P.; Harrison, P.R.; Gasco, M.; Garrone, O.; Crook, T.; Ryan, K.M. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006, 126, 121–134.
- Tasdemir, E.; Maiuri, M.C.; Galluzzi, L.; Vitale, I.; Djavaheri, M.M.; D’Amelio, M.; Criollo, A.; Morselli, E.; Zhu, C.; Harper, F.; et al. Regulation of autophagy by cytoplasmic p53. Nat. Cell Biol. 2008, 10, 676–687.
- Kumar, S.; Cakouros, D. Transcriptional control of the core cell-death machinery. Trends Biochem. Sci. 2004, 29, 193–199.
- Polager, S.; Ofir, M.; Ginsberg, D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene 2008, 27, 4860–4864.
- Lee, I.H.; Cao, L.; Mostoslavsky, R.; Lombard, D.B.; Liu, J.; Bruns, N.E.; Tsokos, M.; Alt, F.W.; Finkel, T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. USA 2008, 105, 3374–3379.
- Sena, L.A.; Chandel, N.S. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 2012, 48, 158–167.
- Scherz, S.R.; Shvets, E.; Fass, E.; Shorer, H.; Gil, L.; Elazar, Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007, 26, 1749–1760.
- Yan, Y.; Finkel, T. Autophagy as a regulator of cardiovascular redox homeostasis. Free Radic. Biol. Med. 2012, 109, 108–113.
- Massey, A.C.; Zhang, C.; Cuervo, A.M. Chaperone-mediated autophagy in aging and disease. Curr. Top. Dev. Biol. 2006, 73, 205–235.
- Pellacani, C.; Costa, L.G. Role of autophagy in environmental neurotoxicity. Environ. Pollut. 2018, 235, 791–805.
- Vidoni, C.; Follo, C.; Savino, M.; Melone, M.A.; Isidoro, C. The Role of Cathepsin D in the Pathogenesis of Human Neurodegenerative Disorders. Med.Res.Rev. 2016, 36, 845–870.
- Priyadarsini, K.I. The chemistry of curcumin: From extraction to therapeutic agent. Molecules 2014, 19, 20091–20112.
- Sharma, O.P. Antioxidant activity of curcumin and related compounds. Biochem. Pharmacol. 1976, 25, 1811–1812.
- Hewlings, S.J.; Kalman, D.S. Curcumin: A Review of Its’ Effects on Human Health. Foods 2017, 6, 92.
- Manson, M.M. Inhibition of survival signalling by dietary polyphenols and indole-3-carbinol. Eur.J. Cancer 2005, 41, 1842–1853.
- Esposito, T.; Schettino, C.; Polverino, P.; Allocca, S.; Adelfi, L.; D’Amico, A.; Capaldo, G.; Varriale, B.; Di Salle, A.; Peluso, G.; et al. Synergistic Interplay between Curcumin and Polyphenol-Rich Foods in the Mediterranean Diet: Therapeutic Prospects for Neurofibromatosis 1 Patients. Nutrients 2017, 9, 783.
- Squillaro, T.; Schettino, C.; Sampaolo, S.; Galderis, I.U.; Di Iorio, G.; Giordano, A.; Melone, M.A.B. Adult-onset brain tumors and neurodegeneration: Are polyphenols protective? J. Cell Physiol. 2017, 233, 3955–3967.
- Wojcik, M.; Krawczyk, M.; Wojcik, P.; Cypryk, K.; Wozniak, L.A. Molecular Mechanisms Underlying Curcumin-Mediated Therapeutic Effects in Type 2 Diabetes and Cancer. Oxid.Med. Cell Longev. 2018, 2018, 9698258.
- Sharma, R.A.; McLelland, H.R.; Hill, K.A.; Ireson, C.R.; Euden, S.A.; Manson, M.M.; Pirmohamed, M.; Marnett, L.J.; Gescher, A.J.; Steward, W.P. Pharmacodynamic and pharmacokinetic study of oral Curcuma extract in patients with colorectal cancer. Clin. Cancer Res. 2001, 7, 1894–1900.
- Garcia, A.M.; Borrelli, L.A.; Rozkalne, A.; Hyman, B.T.; Bacska, I.B.J. Curcumin labels amyloid pathology in vivo, disrupts existing plaques, and partially restores distorted neurites in an Alzheimer mouse model. J. Neurochem. 2007, 102, 1095–1104.
- Maiti, P.; Hall, T.C.; Paladugu, L.; Kolli, N.; Learman, C.; Rossignol, J.; Dunbar, G.L. A comparative study of dietary curcumin, nanocurcumin, and other classical amyloid-binding dyes for labeling and imaging of amyloid plaques in brain tissue of 5×-familial Alzheimer’s disease mice. Histochem. Cell Biol. 2016, 146, 609–625.
- Masuelli, L.; Di Stefano, E.; Fantini, M.; Mattera, R.; Benvenuto, M.; Marzocchella, L.; Sacchetti, P.; Focaccetti, C.; Bernardini, R.; Tresoldi, I.; et al. Resveratrol potentiates the in vitro and in vivo anti-tumoral effects of curcumin in head and neck carcinomas. Oncotarget 2014, 5, 10745–10762.
- Mehanny, M.; Hathout, R.M.; Geneidi, A.S.; Mansour, S. Exploring the use of nanocarrier systems to deliver the magical molecule; Curcumin and its derivatives. J.Control. Release 2016, 225, 1–30.
- Squillaro, T.; Cimini, A.; Peluso, G.; Giordano, A.; Melone, M.A.B. Nano-delivery systems for encapsulation of dietary polyphenols: An experimental approach for neurodegenerative diseases and brain tumors. Biochem. Pharmacol. 2018, 154, 303–317.
- Rajitha, B.; Belalcazar, A.; Nagaraju, G.P.; Shaib, W.L.; Snyder, J.P.; Shoji, M.; Pattnaik, S.; Alam, A.; El-Rayes, B.F. Inhibition of NF-κB translocation by curcumin analogs induces G0/G1 arrest and downregulates thymidylate synthase in colorectal cancer. Cancer Lett. 2016, 373, 227–233.
- Rajitha, B.; Nagaraju, G.P.; Shaib, W.L.; Alese, O.B.; Snyder, J.P.; Shoji, M.; Pattnaik, S.; Alam, A.; El-Rayes, B.F. Novel synthetic curcumin analogs as potent antiangiogenic agents in colorectal cancer. Mol. Carcinog. 2017, 56, 288–299.
- Finicelli, M.; Squillaro, T.; Di Cristo, F.; Di Salle, A.; Melone, M.A.B.; Galderisi, U.; Peluso, G. Metabolic syndrome, Mediterranean diet, and polyphenols: Evidence and perspectives. J. Cell Physiol. 2019, 234, 5807–5826.
- Ye, B.; Wang, Q.; Hu, H.; Shen, Y.; Fan, C.; Chen, P.; Ma, Y.; Wu, H.; Xiang, M. Restoring autophagic flux attenuates cochlear spiral ganglion neuron degeneration by promoting TFEB nuclear translocation via inhibiting MTOR. Autophagy 2019, 15, 998–1016.
- Song, J.X.; Sun, Y.R.; Peluso, I.; Zeng, Y.; Yu, X.; Lu, J.H.; Xu, Z.; Wang, M.Z.; Liu, L.F.; Huang, Y.Y.; et al. A novel curcumin analog binds to and activates TFEB in vitro and in vivo independent of MTOR inhibition. Autophagy 2016, 12, 1372–1389.




