Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | GRACIELA CASTRO-ESCARPULLI | + 1910 word(s) | 1910 | 2021-06-13 06:15:36 | | | |
| 2 | Vicky Zhou | Meta information modification | 1910 | 2021-06-26 11:28:57 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Castro-Escarpulli, G. Aeromonas spp.. Encyclopedia. Available online: https://encyclopedia.pub/entry/11332 (accessed on 08 February 2026).
Castro-Escarpulli G. Aeromonas spp.. Encyclopedia. Available at: https://encyclopedia.pub/entry/11332. Accessed February 08, 2026.
Castro-Escarpulli, Graciela. "Aeromonas spp." Encyclopedia, https://encyclopedia.pub/entry/11332 (accessed February 08, 2026).
Castro-Escarpulli, G. (2021, June 26). Aeromonas spp.. In Encyclopedia. https://encyclopedia.pub/entry/11332
Castro-Escarpulli, Graciela. "Aeromonas spp.." Encyclopedia. Web. 26 June, 2021.
Copy Citation
Aeromonas a Gram-negative bacillus, positive for oxidase and catalase tests, a glucose fermenter, and it is resistant to vibriostatic O/129 (2,4-diamino-6,7-diisopropylpteridine). In humans, it can cause intestinal and extra-intestinal infections. It is important in the medical area, mainly in patients with diarrhea, or with infections in the skin and soft tissue; moreover, it can cause bacteremia, which progresses to sepsis, or endocarditis.
colistin
antimicrobial resistance
1. Introduction
Colistin is a lipopeptide antibiotic from the group of polymyxins. It has a cyclic peptide chain that is linked to a fatty acid. Colistin is used in the medical field, since it is an extended-spectrum antimicrobial, and it is used as a last line of treatment in human infections that are caused by Gram-negative bacilli [1]. Until 2016, resistance to colistin was reported in some genera of bacteria intrinsically and contained in the bacterial genophore, until the presence of a gene called mcr, present in a plasmid that confers resistance to this antimicrobial, was detected in an Escherichia coli strain [2]. After this report, the number of isolates of various origins with mcr genes and a colistin resistance phenotype was increased; in addition, it was found that resistance to this molecule could be transferred horizontally [3][4].
The use of colistin as a treatment for infections increased after the appearance of multidrug resistance phenotypes (MDR) in Gram-negative bacilli and the appearance of carbapenemase-producing enterobacteria type KPC (Klebsiella pneumoniae carbapenemase) or NDM (New Delhi metallo-β-lactamase), in addition to Gram-negative bacilli classified as XDR (extensively drug resistant) that continue to appear, especially in bacteria such as Klebsiella pneumoniae, Pseudomonas aeruginosa, and other Gram-negative bacilli, such as Aeromonas, which is also of medical and veterinary importance and is isolated from environmental samples [5][6].
Aeromonas is a Gram-negative bacillus, positive for oxidase and catalase tests, a glucose fermenter, and it is resistant to vibriostatic O/129 (2,4-diamino-6,7-diisopropylpteridine) [7]. In humans, it can cause intestinal and extra-intestinal infections. It is important in the medical area, mainly in patients with diarrhea, or with infections in the skin and soft tissue; moreover, it can cause bacteremia, which progresses to sepsis, or endocarditis [8][9].
The genus Aeromonas is widely distributed in diverse ecosystems; however, it is a bacterium native to aquatic systems, hence the largest number of isolates are from water. Isolates have been obtained from drinking water, wastewater, bottled water, seawater, and deep and surface water samples. Food isolates have been obtained from vegetables, fruits, pork, poultry, and beef, as well as seafood and fish. In animals, it is considered a pathogen, especially in fish, in which it can cause furunculosis, ulcers, and hemorrhages, among other diseases. This pathogen has also been isolated from infections in rabbits, dogs, cats, chickens, horses, and crustaceans [8][9].
2. Antimicrobial Resistance in Aeromonas
The molecular basis of antimicrobial resistance in Aeromonas spp., has been widely studied, but their importance in the hospital area as a cause of outbreaks is not fully established. Likewise, reports on resistance are varied regarding the origin of isolation and the type of antimicrobials tested in vitro [7]. The Aeromonas resistance profile has not changed significantly; until now, the mechanism of action of inducible chromosomal β-lactamases and carbapenemase expression has been suggested as being the main resistance mechanism for Aeromonas species. Three classes of β-lactamases are recognized in Aeromonas; one of class C cephalosporinase, one of class D penicillinase, and one of class B metallo-β-lactamase (MBL) [9][10].
The occurrence of MDR-type Aeromonas spp., isolates has been increasing. Different authors have suggested that antimicrobial resistance in the clinical setting is closely related to resistance mechanisms detected in environmental isolates [11]. In the genus Aeromonas, the occurrence of MDR strains is equivalent, due to their origin in aquatic environments, which is attributed to the extensive use of antibiotics in aquaculture. Therefore, this environment becomes an ideal setting for the acquisition of these mechanisms of resistance to antimicrobials and other toxic agents [12].
Colistin Resistance in Aeromonas
Since the report in 2016, where it was shown that colistin resistance can be encoded by the mcr genes detected within a plasmid, it was determined that these genes are not only in bacterial genophores but can also be present in mobile genetic elements as plasmids [2]. From this report, attention was paid to the search for and detection of these genes in different bacterial genera, mainly those of medical importance, but also isolated from other sources, such as the environment or animals.In K. pneumoniae, P. aeruginosa, or others, such as Aeromonas genus, mcr gene variants have been detected, and the reports are increasing [13][14].
Colistin resistance in Aeromonas has been reported in several regions of the world, mainly in Europe and Asia. Resistance to this antibiotic has been reported in Latin America in other genera, but not in Aeromonas. The detection of colistin resistance is more common in the clinical area; however, colistin-resistant strains of Aeromonas have been isolated from other origins that have been detected, from which investigations and reports have emerged in the world. The extensive use of antibiotics in aquaculture and in human treatment has led to an increase in the resistance of this genus to antimicrobial drugs [11][12].
The species A. dhakensis, A. hydrophila, A. caviae, and A. veronii are considered the main causes of human infections that can cause infection in wounds, diarrheal syndromes, and other clinical presentations [9]. Commonly, the isolates do not present resistance to antimicrobials; however, MDR isolates have still appeared, and in recent years the report of Aeromonas isolates from clinical samples and from various sources with resistance to colistin has increased [15]. This resistance has been investigated in Aeromonas spp., isolates, by means of disk diffusion test and by minimum inhibitory concentration (MIC), showing the MIC method to be more effective. Induction of colistin resistance in the strains showed an 85% increase after overnight incubation in a tube with Müller–Hinton broth and a 50 µL colistin disk. This result allowed the establishment of a phenotypic marker in the Aeromonas isolates [16].
In an A. veronii isolate from chicken meat, two adjacent genes with colistin resistance markers, called mcr-3.3 and mcr-3-like, were detected in the genophore. The result had 95.2 and 84.19% identity in the nucleotide sequence, when compared to the mcr-3 gene of an E. coli of porcine origin [17].
The evidence of the mcr genes in Aeromonas was demonstrated by a group of scientists who analyzed a total of 6497 strains that were collected in 13 provinces of China between 2016 and 2017. In these samples, the presence of the mcr-3 genes was detected by PCR. The mcr-3 gene was detected in 49 strains only, of which eight strains corresponded to the genus Aeromonas, two A. hydrophila strains, one with mcr-3.8 variant, and one with mcr-3.9 variant, one A. caviae with mcr-3.1 variant, and one A. media with mcr-3.6 variant. Of the four remaining strains, one each were of A. veronii, A. media, and A. caviae, and one was Aeromonas spp., with mcr-3 without variant. All the strains were grouped into a subclade, after the phylogenetic analysis of the sequences of the mcr-3 genes detected in the strains [18].
Another group of researchers found four Aeromonas isolates with the presence of the mcr-3 gene through PCR, while the mcr-1 or mcr-2 genes were not detected. Each of the four isolates with mcr-3 genes presented a different variant each; these presented identities in the amino acid chain were of 95 to 98% compared to the original protein MCR-3. These variants of the protein were designated as MCR-3.6 obtained from the A. allosaccharophila strain isolated from Leuciscus idus, MCR-3.7 for the protein detected in the A. media strain isolated from Meleagris gallopavo, MCR-3.8 for that detected in the A. jandaei strain isolated from a Cyprinus carpio carp, and MCR-3.9 for the protein of the A. hydrophila strain of Cyprinus carpio. The isolate with the mcr-3.9 gene also contained an additional mcr-3.8 gene in the MIC test, with colistin showing an MIC ≥ 128 mg/L higher compared to the other isolates [19].
The reports include a new variant of the mcr-3 gene in A. caviae, also detected in Proteus mirabilis and E. coli that were isolated from a domestic duck. These strains were obtained from sewage samples from free-range ducks, which were raised near a river in the suburban area of Qingdao, Shangdon Province, in China. The presence of the mcr-3 gene was demonstrated in 1 of 15 samples processed in this study. The result was obtained by detection of the mcr gene directly in the sample; the positive sample was seeded in a CHROMagar plate from Biomerieux®, France, to which 2 mg/L of colistin was added. Based on the above, three positive strains were detected for mcr-3 gene. A. caviae 17AC, P. mirabilis 17PM, and E. coli 17EC strains were identified by MALDI-TOF (Matrix-Assisted Laser Desorption/Ionization-Time of Flight) technology, and by 16S rRNA gene sequencing [19]. In another study, the prevalence was determined, complemented by a genetic analysis of the mcr-3 gene in Aeromonas species. These isolates were obtained from human rectal exudates, meat for human consumption, and environmental water samples [20].
The variant mcr-5 gene of colistin resistance was detected in an A. hydrophila strain isolated from a fecal sample from a backyard pig. In this case, the mcr-5 gene was detected in a plasmid with 7915 base pairs (bp) named pI064-2. Additionally, they analyzed the possibility of transforming the A. hydrophila strains susceptible to colistin into a resistant strain [21].
Various mechanisms of resistance to colistin, in addition to the mechanism mediated by mcr genes, have been described in some bacteria, including P. aeruginosa, A. baumannii, members of the Enterobacteriaceae family, such as E. coli, Salmonella spp., and K. pneumoniae they have an acquired resistance against colistin. However, the possibility of the appearance of strains resistant to this antibiotic should be monitored, due to the presence of mutations, new mechanisms, or adaptations [22].
The protein generated by the mcr genes confers resistance to polymyxins; this protein, called MCR, from the inner membrane, adds a molecule of phosphoethanolamine (PEA) to lipid A of lipopolysaccharide (Kdo2-PEA-Lipid A), synthesized by the binding of uridine diphosphate (UDP) and N-acetyl glucosamine (GlcNac) mediated by Lpx proteins (C, D, H, B, K, L, M). The product binds to 2-keto deoxioctanate acid (Kdo2), which is transported by the MsbA protein to the inner membrane, where the PEA molecule is added. The Lpt protein complex (ABCFD/DE) transports the modified LPS that generates resistance to colistin, since the negative charge with which it interacts was modified (Figure 1) [23][24][25].
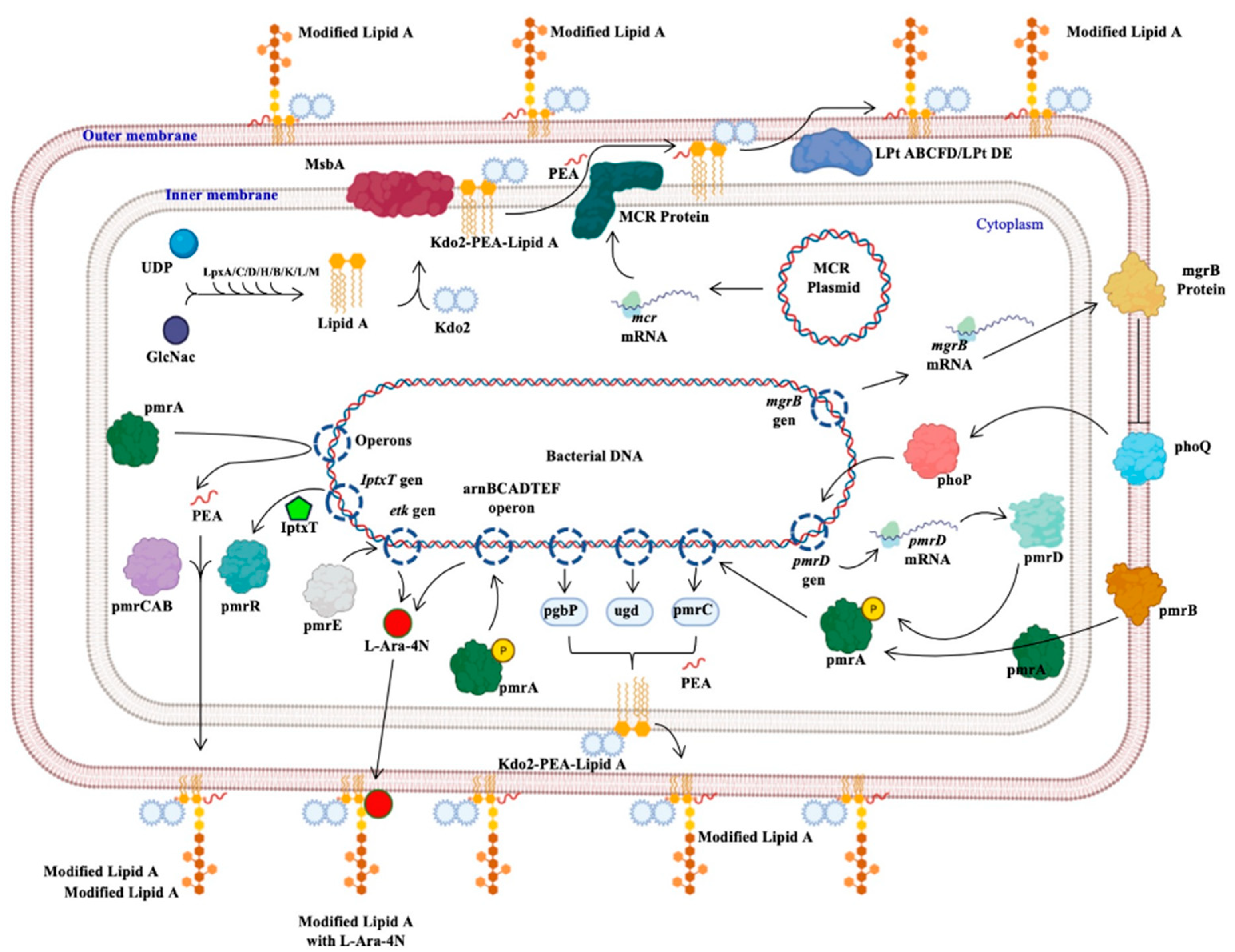 Figure 1. Bacterial mechanisms of resistance to colistin. The different mechanisms of resistance to colistin described in Gram-negative bacilli and other bacteria are presented: the mechanism mediated by the mcr genes, those regulated by the phoP and phoQ proteins, others regulated by the pmr proteins or by the iptxT and etk genes, and operons that encode activation proteins of pmr proteins [22][23][24][25]. Generated from this work.
Figure 1. Bacterial mechanisms of resistance to colistin. The different mechanisms of resistance to colistin described in Gram-negative bacilli and other bacteria are presented: the mechanism mediated by the mcr genes, those regulated by the phoP and phoQ proteins, others regulated by the pmr proteins or by the iptxT and etk genes, and operons that encode activation proteins of pmr proteins [22][23][24][25]. Generated from this work.3. Conclusions
The genus Aeromonas is a bacterium widely distributed in the environment. Its presence in various ecosystems and ability to cause infections in animals and humans makes it a bacterium of interest in the study related to the appearance of strains resistant to antimicrobials. The presence of antimicrobial-resistant isolates from the first line of care, and those of the last alternative, such as colistin, detected in the clinical area, in animals or in the environment, demonstrates that the resistance comes from the exposure of bacteria to antimicrobials in clinical care, but it is also suggested that resistance originates in the environment. In addition, it is important to study Aeromonas isolates with colistin resistance from different sources, and also with the detection of the mcr genes.
References
- Upert, G.; Luther, A.; Obrecht, D.; Ermert, P. Emerging peptide antibiotics with therapeutic potential. Med. Drug Discov. 2021, 9, 100078.
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Yu, L.F. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168.
- Medina, J.; Paciel, D.; Noceti, O.; Rieppi, G. Actualización acerca de colistina (polimixina E): Aspectos clínicos, PK/PD y equivalencias. Rev. Méd. Uru. 2017, 33, 79–114.
- Karaiskos, I.; Souli, M.; Galani, I.; Giamarellou, H. Colistin: Still a lifesaver for the 21st century? Expert Opin. Drug. Met. 2017, 13, 59–71.
- Anandan, S.; Gopi, R.; Ragupathi, N.K.D.; Sethuvel, D.P.M.; Gunasekaran, P.; Walia, K.; Veeraraghavan, B. First report of blaOXA-181-mediated carbapenem resistance in Aeromonas caviae in association with pKP3-A: Threat for rapid dissemination. J. Glob. Antimicrob. Resist. 2017, 10, 310–314.
- Jiménez-Pearson, M.A.; Galas, M.; Corso, A.; Hormazábal, J.C.; Duarte-Valderrama, C.; Salgado-Marcano, N.; Melano, R.G. Consenso latinoamericano para definir, categorizar y notificar patógenos multirresistentes, con resistencia extendida o panresistentes. Rev. Panam. Salud Púb. 2019, 43, e65.
- Janda, J.M.; Abbott, S.L. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010, 23, 35–73.
- Figueras, M.J.; Beaz-Hidalgo, R. Aeromonas infections in humans. In Aeromonas, 1st ed.; Graf, J., Ed.; Caister Academic Press: Pole, UK, 2015; Chapter 4; pp. 65–108.
- Fernández-Bravo, A.; Figueras, M.J. An Update on the Genus Aeromonas: Taxonomy, Epidemiology, and Pathogenicity. Microorganisms 2020, 8, 129.
- Tekedar, H.C.; Kumru, S.; Blom, J.; Perkins, A.D.; Griffin, M.J.; Abdelhamed, H.; Karsi, A.; Lawrence, M.L. Comparative genomics of Aeromonas veronii: Identification of a pathotype impacting aquaculture globally. PLoS ONE 2019, 14, e0221018.
- Esteve, C.; Alcaide, E.; Giménez, M.J. Multidrug-resistant (MDR) Aeromonas recovered from the metropolitan area of Valencia (Spain): Diseases spectrum and prevalence in the environment. Eur. J. Clin. Microbiol. 2015, 34, 137–145.
- Zhou, Y.; Yu, L.; Nan, Z.; Zhang, P.; Kan, B.; Yan, D.; Su, J. Taxonomy, virulence genes and antimicrobial resistance of Aeromonas isolated from extra-intestinal and intestinal infections. BMC Infect. Dis. 2019, 19, 158.
- Tansarli, G.S.; Papaparaskevas, J.; Balaska, M.; Samarkos, M.; Pantazatou, A.; Markogiannakis, A.; Daikos, G.L. Colistin resistance in carbapenemase-producing Klebsiella pneumoniae bloodstream isolates: Evolution over 15 years and temporal association with colistin use by time series analysis. Int. J. Antimicrob. Agents 2018, 52, 397–403.
- Jorgensen, J.H.; Hindler, J.F.; Reller, L.B.; Weinstein, M.P. New consensus guidelines from the Clinical and Laboratory Standards Institute for antimicrobial susceptibility testing of infrequently isolated or fastidious bacteria. Clin. Infect. Dis. 2007, 44, 280–286.
- Bravo-Fariñas, L.; Cabrera-Rodríguez, L.E.; Margarita-Ramírez, M.; Llop-Hernández, A.; Verdecía-Pérez, J.; Borrego-Hernández, G.; Fernández-Abreu, A. Resistencia antimicrobiana en cepas de Aeromonas spp. aisladas de pacientes con bacteriemia. Rev. Biomédica 2007, 18, 176–181.
- Fosse, T.; Giraud-Morin, C.; Madinier, I. Induced colistin resistance as an identifying marker for Aeromonas phenospecies groups. Lett. Appl. Microbiol. 2003, 36, 25–29.
- Ling, Z.; Yin, W.; Li, H.; Zhang, Q.; Wang, X.; Wang, Z.; Shen, J. Chromosome-mediated mcr-3 variants in Aeromonas veronii from chicken meat. Antimicrob. Agents Chemother. 2017, 61, e01272-17.
- Xu, Y.; Zhong, L.L.; Srinivas, S.; Sun, J.; Huang, M.; Paterson, D.L.; Lei, S.; Lin, J.; Li, X.; Tang, Z.; et al. Spread of MCR-3 colistin resistance in China: An epidemiological, genomic and mechanistic study. EbioMedicine 2018, 34, 139–157.
- Eichhorn, I.; Feudi, C.; Wang, Y.; Kaspar, H.; Feßler, A.T.; Lübke-Becker, A.; Michael, G.B.; Shen, J.; Schwarz, S. Identification of novel variants of the colistin resistance gene mcr-3 in Aeromonas spp. from the national resistance monitoring programme GE RM-Vet and from diagnostic submissions. J. Antimicrob. Chemother. 2018, 73, 1217–1221.
- Shen, Y.; Xu, C.; Sun, Q.; Schwarz, S.; Ou, Y.; Yang, L.; Zhang, R. Prevalence and genetic analysis of mcr-3-positive Aeromonas species from humans, retail meat, and environmental water samples. Antimicrob. Agents Chemother. 2018, 62, e00404-18.
- Ma, S.; Sun, C.; Hulth, A.; Li, J.; Nilsson, L.E.; Zhou, Y.; Wang, Y. Mobile colistin resistance gene mcr-5 in porcine Aeromonas hydrophila. J. Antimicrob. Chemother. 2018, 73, 1777–1780.
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643.
- Aghapour, Z.; Gholizadeh, P.; Ganbarov, K.; Bialvaei, A.Z.; Mahmood, S.S.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Yousefi, B.; Kafil, H.S. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect. Drug Resist. 2019, 12, 965–975.
- Trebosc, V.; Gartenmann, S.; Tötzl, M.; Lucchini, V.; Schellhorn, B.; Pieren, M.; Lociuro, S.; Gitzinger, M.; Tigges, M.; Bumann, D.; et al. Dissecting Colistin Resistance Mechanisms in Extensively Drug-Resistant Acinetobacter baumannii Clinical Isolates. mBio 2019, 10, e01083-19.
- Venter, H.; Henningsen, M.L.; Begg, S.L. Antimicrobial resistance in healthcare, agriculture and the environment: The biochemistry behind the headlines. Essays Biochem. 2017, 61, 1–10.
More
Information
Subjects:
Microbiology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.2K
Revisions:
2 times
(View History)
Update Date:
26 Jun 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




