Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Feng, J. Zuotin-Related Factor 1. Encyclopedia. Available online: https://encyclopedia.pub/entry/13286 (accessed on 08 February 2026).
Feng J. Zuotin-Related Factor 1. Encyclopedia. Available at: https://encyclopedia.pub/entry/13286. Accessed February 08, 2026.
Feng, Jing. "Zuotin-Related Factor 1" Encyclopedia, https://encyclopedia.pub/entry/13286 (accessed February 08, 2026).
Feng, J. (2021, August 18). Zuotin-Related Factor 1. In Encyclopedia. https://encyclopedia.pub/entry/13286
Feng, Jing. "Zuotin-Related Factor 1." Encyclopedia. Web. 18 August, 2021.
Copy Citation
Recently, Zuotin-related factor 1 (ZRF1), an epigenetic regulator, was found to be involved in transcriptional regulation. In animals and humans, ZRF1 specifically binds to monoubiquitinated histone H2A through a ubiquitin-binding domain and derepresses Polycomb target genes at the beginning of cellular differentiation. In addition, ZRF1 can work as a tumor suppressor. According to bioinformatics analysis, ZRF1 homologs are widely found in plants.
ZRF1
Arabidopsis thaliana
stem cell maintenance
embryo development
monoubiquitination
1. Introduction
Epigenetics is the study of inherited changes in phenotype (appearance) or gene expression caused by mechanisms other than the changes in the underlying DNA sequence; hence, the name epi- (Greek: επί- over, above) genetics. Epigenetic regulatory mechanisms comprise histone post-translational modifications, histone variants, DNA methylation, and ATP-dependent chromatin remodeling, which are essential for controlling patterns of gene expression during the cell cycle, development, and in response to environmental stimuli. More importantly, epigenetic changes may be reversed [1][2]. DNA methylation has been generally considered a heritable chromatin modification mark that plays important roles in maintaining chromosome stability, regulating transposon silencing, and regulating gene expression [3][4]. Histone post-translational modifications include methylation, acetylation, phosphorylation, ubiquitination, and sumoylation; they are dynamic changes and play key roles in regulating chromatin structure and gene expression, mainly by changing chromatin compaction and shaping the three-dimensional topology of the genome [5][6][7].
Polycomb Group (PcG) proteins are evolutionarily conserved transcriptional regulators, which are essential for the control of development and cell differentiation in plants and animals. Polycomb-mediated transcriptional regulation is mediated by two major multisubunit complexes: Polycomb Repressive Complex 2, which catalyzes histone H3 lysine 27 trimethylation (H3K27me3) on target chromatin, and PRC1, which not only recognizes and binds H3K27me3, but also catalyzes the mono-ubiquitination of histone H2A lysine 119 (H2AK119ub1), resulting in a stable silencing chromatin state [8].
Zuotin-related factor 1 (ZRF1) (also known as DNAJC2, MIDA1, Mpp11, and GlsA) has been characterized as an epigenetic factor involved in transcriptional regulation. In most species, ZRF1 protein is evolutionarily conserved. The N-terminal of all homologs of ZRF1 has a zuotin domain, which is composed of a DnaJ domain and a potential ubiquitin-binding domain (UBD). The C-terminal contains tandem repeats of SANT (Swi3, Ada2, NcoR1 and TFIIIB) domain, but only exists in higher eukaryotes [9][10]. It has been reported that human ZRF1 protein can specifically recognizes and bind to monoubiquitinated histone H2A through UBD, and activates polycomb-repressed target genes through the E3 ubiquitin ligase activity of Ring1B at the onset of cellular differentiation (e.g., stem cell differentiation, the neuronal lineage, and mesodermal lineage) [10][11][12][13]. In mice, based on the CRISPR/Cas9 approach, it was first suggested that DNAJC2 was necessary during early mouse embryogenesis [12]. Moreover, the nuclear epigenetic factor ZRF1 participates in senescence and is involved in certain cancers [14][15][16][17]. Research evidence suggests that ZRF1 can be phosphorylated on Ser47 residues by S6 kinases 1 and 2 (S6Ks) and participates in a senescence programme by regulating the cell cycle inhibitor p16 [18]. It is very interesting that ZRF1 has a dual role. Not only does it work as a tumor suppressor, but it can also induce carcinogenesis depending on the cellular context [14][19]. In addition, ZRF1, a switch protein, remodels chromatin-bound E3 ligases during lesion recognition [20]. Under ultraviolet radiation, the endonuclease DICER and ZRF1 are required for chromatin decondensation. DICER was recruited into chromatin in a ZRF1-mediated manner. H2A ubiquitin-binding protein ZRF1 interacts with DICER to affect chromatin conformation through chromatin remodeling factors PARP1 [21].
Over the past several years, the functions of ZRF1 in animals and humans have been identified and have become a new research hotspot. In most species, ZRF1 is evolutionarily conserved [9][22]. According to bioinformatics analysis, ZRF1 homologs are widely found in plants (e.g., A. thaliana, Lilium longiflorum, Alstroemeria aurea, Volvox carteri), and LlglsA (L. longiflorum gonidialess A) was identified as the first higher plant homolog of glsA/ZRF1 [22][23][24][25]. However, current studies of ZRF1 in plants are limited, and few in-depth studies of its functions have been reported.
2. ZRF1 Orthologs and the Domain Structure in Arabidopsis
In Arabidopsis, two ZRF1 proteins (AtZRF1a and AtZRF1b) have been identified, and they share 81% sequence identity. AtZRF1a gene, encoding a 74 kDa protein consisting of 647 amino acids, is located on chromosome 3 (At3g11450). AtZRF1b gene, encoding a 75.6 kDa protein composed of 663 amino acids, is located on chromosome 5 (At5g06110). There are no introns within the coding region of both AtZRF1a and AtZRF1b [22]. Both AtZRF1a and AtZRF1b are broadly expressed in various plant organs/tissues, and the single loss-of-function mutants atzrf1a and atzrf1b showed normal phenotypes, while the double mutants atzrf1a atzrf1b have a pleiotropic phenotype [22]. Taken together, the data demonstrate that AtZRF1a and AtZRF1b have redundant functions [22].
The ZRF1 protein structure is highly evolutionarily conserved, and several domains have been annotated [9][14]. Similar to human ZRF1, AtZRF1a and AtZRF1b also contain two major recognizable types of structural domains, namely, the Zuotin domain (Z-DNA binding domain) and SANT (Swi3, Ada2, NcoR1, and TFIIIB) domains (Figure 1 and Figure 2). Zuotin contains a highly conserved DnaJ domain (J-domain), which is similar to the mammalian HSP-40 (heat shock protein 40) chaperone, mediating interactions with heat shock proteins (Hsp70). The green alga V. carteri ZUO1/ZRF ortholog Gonidialess A (GlsA) can bind to the corresponding Hsp70 chaperones, and this interaction is required for asymmetric cell division [26][27]. Zuotin also contains a ubiquitin-binding domain (UBD) domain/M region, which is adjacent to the C terminus of the J-domain. Human MPP11/ZRF1 can bind to the H2Aub epigenetic marker and replace Polycomb repressive complex 1 (PRC1) through the UBD domain [10]. Interestingly, AtZRF1b can bind ubiquitin and can pulldown H2Aub1 via the UBD domain; the loss of AtZRF1a/b does not cause an increase in H2Aub1 but rather results in a very slightly reduced global level of H2Aub1 in atzrf1a-1 atzrf1b-1 mutant plants [22]. Unfortunately, there is no relevant experimental evidence for the interaction between ZRF1a and H2Aub. Except for fungal proteins, all known ZRF1 orthologs contain the tandem repeat of SANT domains at the C terminus. SANT domain proteins can recruit histone acetylases or deacetylases to modify histone tails [28][29][30], but their specific functions remain to be addressed in plants.
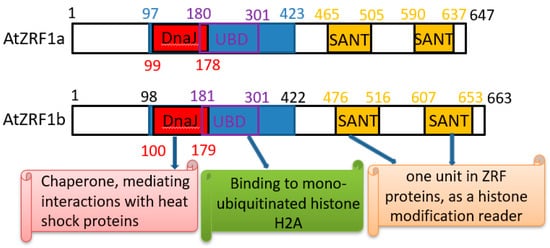
Figure 1. Schematic diagram of Arabidopsis ZRF1a/b for conserved functional domain organization.
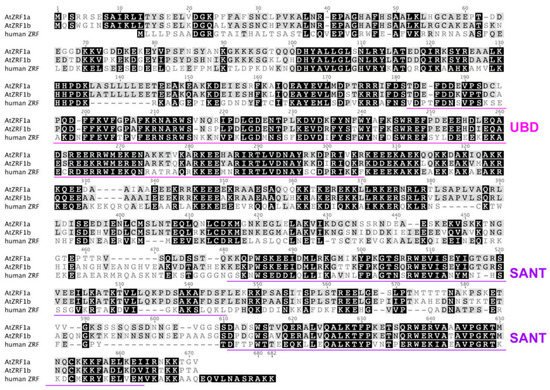
Figure 2. Sequence alignment of AtZRF1a and AtZRF1b together with the human ZRF1. The alignment was generated using the CLUSTALW program. The UBD and SANT domains are indicated [22].
3. ZRF1 Is Required in Seed/Embryo Development in Arabidopsis
A seed consists of a plant embryo and an endosperm enclosed by a seed coat. Several studies have shown that PRC1 is involved in seed development and seedling growth [31][32][33]. The double mutants Atring1a Atring1b, as well as Atbmi1a Atbmi1b, exhibit derepression of embryonic traits in somatic plant tissues, due to the lack of AtRING1 or AtBMI1, the key catalytic factors of PRC1. For example, ectopic callus formed in cotyledon, leaf or SAM region of mutant plants [31][32]. CURLY LEAF (CLF), EMBRYONIC FLOWER2 (EMF2), VERNALIZATION2 (VRN2) and SWINGER (SWN) are PRC2 components, the clf swn and emf2 vrn2 mutants also exhibit ectopic cell dedifferentiation and somatic embryogenesis [34]. Investigation of the triple mutants Atring1a Atring1b clf revealed that the expression of embryo regulatory genes was increased [32][35]. These genetic data suggest that PRC1 and PRC2 jointly regulate embryonic regulatory genes to prevent somatic dedifferentiation.
Compared with wild-type seeds, the seeds of atglsa1-3 atglsa2-1 double mutants are shriveled, wrinkled, distorted, and small [23]. In atzrf1a-1 atzrf1b-1 and atzrf1a-2 atzrf1b-1 double mutants, plants are poorly fertile and seed abortion is observed. Based on reciprocal crosses of heterozygous mutant plants with wild-type Col-0 plants, male and female transmissions were impaired. Some seedlings (about 35%) exhibited a single cotyledon, asymmetrical cotyledons, and fleshy narrow cotyledonous phenotypes, which display some defects similar to Atbmi1aAtbmi1b and Atring1aAtring1b [22]. Through electron microscopy, severe defects in all stages of embryo development were observed, such as at the pre-globular, octant stage, unorganized division, and a lack of suspensor development; at the globular stage, failure of hypophysis development [23]. These phenomena suggest that ZRF1a/b plays a very important role in embryo development. In humans/mice, PRC1 and PRC2 are epigenetic repressors required for proper embryonic stem cells differentiation and embryo development [36]. ZRF1 can replace PRC1 from Chromatin by competing for H2Aub1 markers, thus inducing de-repression of neural genes [10][11]. However, in plants, the relationship between ZRF1 and PRC1/PRC2 in regulating embryonic development is not clear.
Seed germination is the first step in plant postembryonic growth, which is regulated by various pathways. The primary energy for seed germination in higher plants comes from seed storage products. The expression of seed developmental genes (e.g., ABI3 (ABA INSENSI-TIVE 3), CRA1 (CRUCIFERIN1), CHO1 (CHOTTO1)) should be silenced from seed germination to the whole plant growth and development process [37][38][39]. Under standard growth conditions, the double mutants atzrf1a-1 atzrf1b-1 and atzrf1a-2 atzrf1b-1 showed delayed seed germination. ABI3, CRU1/CRA1, CRU3, and CHO1/AIL5, were up-regulated in atzrf1a atzrf1b mutants as well as in atbmi1a atbmi1b mutant seedlings. Further, within the ABI3, CRU3, and AIL5 gene loci, both H2Aub1 and H3K27me3 levels were strongly reduced. Previous studies have shown that de-suppression of ABI3, CRU3, and AIL5 expression is associated with decreased levels of H2Aub1 and H3K27me3 in atbmi1a atbmi1b mutants [22][33]. These results indicate that ZRF1a/b is required to enhance H2Aub1 and H3K27me3 levels for suppressing seed developmental genes in seedlings, similar to AtBMI1a/b. Since ZRF1 is associated with H2Aub1 de-deposition in animals, it is suspected that the complementary mechanisms of the interaction between AtZRF1A/B-H2Aub1 and AtZRF1A/B-PRC1/PRC2 may be involved in the deposition of H2Aub1 and H3k27me3, thus effectively silencing seed development genes and promoting plant vegetative growth. However, further experiments are needed to test this hypothesis.
References
- avalli, G.; Heard, E. Advances in epigenetics link genetics to the environment and disease. Nature 2019, 571, 489–499.
- Inbar-Feigenberg, M.; Choufani, S.; Butcher, D.T.; Roifman, M.; Weksberg, R. Basic concepts of epigenetics. Fertil. Steril. 2013, 99, 607–615.
- He, X.-J.; Chen, T.; Zhu, J.-K. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011, 21, 442–465.
- Tariq, M.; Paszkowski, J. DNA and histone methylation in plants. Trends Genet. 2004, 20, 244–251.
- Feng, J.; Shen, W.-H. Dynamic regulation and function of histone monoubiquitination in plants. Front. Plant Sci. 2014, 5.
- Mozgova, I.; Hennig, L. The Polycomb Group Protein Regulatory Network. Annu. Rev. Plant Biol. 2015, 66, 269–296.
- Pfluger, J.; Wagner, D. Histone modifications and dynamic regulation of genome accessibility in plants. Curr. Opin. Plant Biol. 2007, 10, 645–652.
- Schuettengruber, B.; Chourrout, D.; Vervoort, M.; Leblanc, B.; Cavalli, G. Genome Regulation by Polycomb and Trithorax Proteins. Cell 2007, 128, 735–745.
- Chen, D.-H.; Huang, Y.; Liu, C.; Ruan, Y.; Shen, W.-H. Functional conservation and divergence of J-domain-containing ZUO1/ZRF orthologs throughout evolution. Planta 2014, 239, 1159–1173.
- Richly, H.; Rocha-Viegas, L.; Ribeiro, J.D.; Demajo, S.; Gundem, G.; Lopez-Bigas, N.; Nakagawa, T.; Rospert, S.; Ito, T.; Di Croce, L. Transcriptional activation of polycomb-repressed genes by ZRF1. Nat. Cell Biol. 2010, 468, 1124–1128.
- Aloia, L.; Di Stefano, B.; Sessa, A.; Morey, L.; Santanach, A.; Gutierrez, A.; Cozzuto, L.; Benitah, S.A.; Graf, T.; Broccoli, V.; et al. Zrf1 is required to establish and maintain neural progenitor identity. Genes Dev. 2014, 28, 182–197.
- Helary, L.; Castille, J.; Passet, B.; Vaiman, A.; Beauvallet, C.; Jaffrezic, F.; Charles, M.; Tamzini, M.; Baraige, F.; Letheule, M.; et al. DNAJC2 is required for mouse early embryonic development. Biochem. Biophys. Res. Commun. 2019, 516, 258–263.
- Ribeiro, J.D.; Morey, L.; Mas, A.; Gutierrez, A.; Luis, N.M.; Mejetta, S.; Richly, H.; Benitah, S.A.; Keyes, W.M.; Di Croce, L. ZRF1 controls oncogene-induced senescence through the INK4-ARF locus. Oncogene 2013, 32, 2161–2168.
- Aloia, L.; Demajo, S.; Di Croce, L. ZRF1: A novel epigenetic regulator of stem cell identity and cancer. Cell Cycle 2015, 14, 510–515.
- Imamura, T.; Komatsu, S.; Ichikawa, D.; Miyamae, M.; Okajima, W.; Ohashi, T.; Kiuchi, J.; Nishibeppu, K.; Kosuga, T.; Konishi, H.; et al. Overexpression of ZRF1 is related to tumor malignant potential and a poor outcome of gastric carcinoma. Carcinogenesis 2018, 39, 263–271.
- Kaymak, A.; Sayols, S.; Papadopoulou, T.; Richly, H. Role for the transcriptional activator ZRF1 in early metastatic events in breast cancer progression and endocrine resistance. Oncotarget 2018, 9, 28666–28690.
- Kaymak, A.; Richly, H. Zrf1 controls mesoderm lineage genes and cardiomyocyte differentiation. Cell Cycle 2016, 15, 3306–3317.
- Barilari, M.; Bonfils, G.; Treins, C.; Koka, V.; De Villeneuve, D.; Fabrega, S.; Pende, M. ZRF 1 is a novel S6 kinase substrate that drives the senescence programme. EMBO J. 2017, 36, 736–750.
- Demajo, S.; Uribesalgo, I.; Gutiérrez, A.; Ballare, C.; Capdevila, S.; Roth, M.; Zuber, J.; Martín-Caballero, J.; Di Croce, L. ZRF1 controls the retinoic acid pathway and regulates leukemogenic potential in acute myeloid leukemia. Oncogene 2014, 33, 5501–5510.
- Gracheva, E.; Chitale, S.; Wilhelm, T.; Rapp, A.; Byrne, J.; Stadler, J.; Medina, R.; Cardoso, M.C.; Richly, H. ZRF1 mediates remodeling of E3 ligases at DNA lesion sites during nucleotide excision repair. J. Cell Biol. 2016, 213, 185–200.
- Chitale, S.; Richly, H. DICER and ZRF1 contribute to chromatin decondensation during nucleotide excision repair. Nucleic Acids Res. 2017, 45, 5901–5912.
- Feng, J.; Chen, D.; Berr, A.; Shen, W.-H. ZRF1 Chromatin Regulators Have Polycomb Silencing and Independent Roles in Development. Plant Physiol. 2016, 172, 1746–1759.
- Guzmán-López, J.A.; Abraham, J.; Lozano-Sotomayor, P.; de Folter, S.; Simpson, J. Arabidopsis thaliana gonidialess A/Zuotin related factors (GlsA/ZRF) are essential for maintenance of meristem integrity. Plant Mol. Biol. 2016, 91, 37–51.
- Igawa, T.; Hoshino, Y.; Yanagawa, Y. Isolation and characterization of the plant glsA promoter from Alstroemeria. Plant Biol. 2009, 11, 878–885.
- Mori, T.; Kuroiwa, H.; Higashiyama, T.; Kuroiwa, T. Identification of higher plant GlsA, a putative morphogenesis factor of gametic cells. Biochem. Biophys. Res. Commun. 2003, 306, 564–569.
- Cheng, Q.; Pappas, V.; Hallmann, A.; Miller, S.M. Hsp70A and GlsA interact as partner chaperones to regulate asymmetric division in Volvox. Dev. Biol. 2005, 286, 537–548.
- Miller, S.; Kirk, D. glsA, a Volvox gene required for asymmetric division and germ cell specification, encodes a chaperone-like protein. Development 1999, 126, 649–658.
- Badri, K.R.; Zhou, Y.; Dhru, U.; Aramgam, S.; Schuger, L. Effects of the SANT Domain of Tension-Induced/Inhibited Proteins (TIPs), Novel Partners of the Histone Acetyltransferase p300, on p300 Activity and TIP-6-Induced Adipogenesis. Mol. Cell. Biol. 2008, 28, 6358–6372.
- Boyer, L.A.; Latek, R.R.; Peterson, C.L. The SANT domain: A unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 2004, 5, 158–163.
- Mo, X.; Kowenz-Leutz, E.; Laumonnier, Y.; Xu, H.; Leutz, A. Histone H3 tail positioning and acetylation by the c-Myb but not the v-Myb DNA-binding SANT domain. Genes Dev. 2005, 19, 2447–2457.
- Bratzel, F.; López-Torrejón, G.; Koch, M.; del Pozo, J.C.; Calonje, M. Keeping Cell Identity in Arabidopsis Requires PRC1 RING-Finger Homologs that Catalyze H2A Monoubiquitination. Curr. Biol. 2010, 20, 1853–1859.
- Chen, D.; Molitor, A.; Liu, C.; Shen, W.-H. The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res. 2010, 20, 1332–1344.
- Molitor, A.M.; Bu, Z.; Yu, Y.; Shen, W.-H. Arabidopsis AL PHD-PRC1 Complexes Promote Seed Germination through H3K4me3-to-H3K27me3 Chromatin State Switch in Repression of Seed Developmental Genes. PLoS Genet. 2014, 10, e1004091.
- Chanvivattana, Y.; Bishopp, A.; Schubert, D.; Stock, C.; Moon, Y.-H.; Sung, Z.R.; Goodrich, J. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 2004, 131, 5263–5276.
- Xu, L.; Shen, W.-H. Polycomb Silencing of KNOX Genes Confines Shoot Stem Cell Niches in Arabidopsis. Curr. Biol. 2008, 18, 1966–1971.
- Aloia, L.; Di Stefano, B.; Di Croce, L. Polycomb complexes in stem cells and embryonic development. Development 2013, 140, 2525–2534.
- Dekkers, B.J.; He, H.; Hanson, J.; Willems, L.A.; Jamar, D.C.; Cueff, G.; Rajjou, L.; Hilhorst, H.; Bentsink, L. The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. Plant J. 2016, 85, 451–465.
- Gliwicka, M.; Nowak, K.; Cieśla, E.; Gaj, M.D. Expression of seed storage product genes (CRA1 and OLEO4) in embryogenic cultures of somatic tissues of Arabidopsis. Plant Cell Tissue Organ Cult. 2012, 109, 235–245.
- Yano, R.; Kanno, Y.; Jikumaru, Y.; Nakabayashi, K.; Kamiya, Y.; Nambara, E. CHOTTO1, a Putative Double APETALA2 Repeat Transcription Factor, Is Involved in Abscisic Acid-Mediated Repression of Gibberellin Biosynthesis during Seed Germination in Arabidopsis. Plant Physiol. 2009, 151, 641–654.
More
Information
Subjects:
Genetics & Heredity
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Revisions:
2 times
(View History)
Update Date:
18 Aug 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




